Abstract
Bitter rot caused by the fungal genus Colletotrichum is a well-known, common disease of apple and causes significant yield loss. In 2013, six fungal strains were isolated from Fuji apple fruits exhibiting symptoms of bitter rot from Andong, Korea. These strains were identified as Colletotrichum fructicola and C. siamense based on morphological characteristics and multilocus sequence analysis of the internal transcribed spacer rDNA, actin, calmodulin, chitin synthase, and glyceraldehyde-3-phosphate dehydrogenase Pathogenicity tests confirmed the involvement of C. fructicola and C. siamense in the development of disease symptoms on apple fruits. This is the first report of C. fructicola and C. siamense causing bitter rot on apple fruit in Korea.
1. Introduction
Bitter rot caused by fungi in the genus Colletotrichum is one of the most common diseases of apple fruit and causes significant yield loss of apple crop worldwide [Citation1]. Colletotrichum gloeosporioides and C. acutatum are the two most common species causing bitter rot on apples [Citation2]. It is difficult to identify Collectitrichum to species based only on morphology, due to the morphological similarity at different growth conditions [Citation3]. Recently, Colletotrichum was studied based on morphology and multilocus phylogenetic analysis, grouping the 189 recognized species into 11 species complexes [Citation4]. Additional studies described several new species from different hosts in the C. acutatum and C. gloeosporioides complexes [Citation5,Citation6]. Of the described species, five have been reported to cause bitter rot of apple fruit: C. fioriniae and C. nymphaeae (C. acutatum species complex) and C. siamense, C. theobromicola, and C. fructicola (C. gloeosporioides species complex) [Citation1].
Apple (Malus pumila Mill.) is grown in worldwide. In Korea, the total area of apple cultivation is approximately 31,620 ha and is gradually increasing (KOSTAT, Statistics Korea, http://kostat.go.kr/portal/eng/). To date, about 40 pathogens have known associations with apple in Korea, including anthracnose, white rot, Alternaria leaf spot, Marssonina blotch, and bacterial shoot blight [Citation7]. Two species of Colletotrichum (C. gloeosporioides and C. acutatum) were reported to be associated with apple anthracnose disease in Korea [Citation7]. Bitter rot is a serious disease of apple fruit in Korea and causes significant yield loss [Citation8]. C. gloeosporioides and C. acutatum were previously identified as causative agents bitter rot of apple in Korea [Citation7,Citation9]. These studies were based on morphological features and sequence analysis of a single gene (ITS or β-tubulin) – data that has now been shown to be unreliable (morphological plasticity, low resolution of DNA) for species identification. The aim of the present study was to identify the causative agent of bitter rot on apple fruit from Andong, Korea. We identify six unknown fungal strains associated with bitter rot based on morphological features and multilocus sequence analyses of the internal transcribed spacer (ITS) rDNA, actin (ACT), calmodulin (CAL), chitin synthase (CHS-1), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and provide verification with pathogenicity tests.
2. Materials and methods
2.1. Sampling and isolation
Fungal isolates were collected from Fuji apple fruit with symptoms of bitter rot collected from orchards in Andong, Korea during 2013. The fruit tissues were surface-sterilized by dipping them in 0.1% NaOCl solution for 1 min, rinsed three times with sterilized distilled water, and placed on water agar at 25 °C for two days. The margins of each hyphae growing from the tissues were transferred to potato dextrose agar (PDA, Difco, Becton Dickinson) plates and incubated at 25 °C for one week. Fungal isolates were maintained in 20% glycerol at –80 °C for identification and pathogenicity assay at the Chungcheongnam-do Agricultural Research and Extension Services (CNARES).
2.2. DNA extraction, PCR, sequencing and phylogenetic analyses
Genomic DNA was extracted directly from mycelia of fungal strains using a modified cetyltrimethylammonium bromide (CTAB) extraction protocol [Citation10]. PCR amplifications of ITS, ACT, CAL, CHS-1, and GAPDH were performed in a C1000 Thermal Cycler (Bio-Rad, Hercules, CA, USA) using previously described methods [Citation6]. DNA sequencing was performed at Macrogen (Seoul, Korea), using an ABI PRISM 3730XL Analyzer (Life Technologies, Gaithersburg, MD, USA). Sequences were proofread and edited using MEGA ver. 5.0 [Citation11] and aligned with reference sequences of type strains downloaded from GenBank using the default settings of MAFFT v7 [Citation12]. Maximum likelihood phylogenetic analyses were performed using RAxML [Citation13] for an ITS only dataset for initial identification, followed by the concatenated data set (ITS, ACT, CAL, CHS-1, and GAPDH) for species identification. Analyses were run with the GTR + G model of evolution and 1000 bootstrap replicates. All sequences were deposited in GenBank ().
Table 1. Information and GenBank accession numbers of Colletotrichum strains isolated this study.
Table 2. Comparison of the morphological characteristics of the strain used in this study with those of previously reported C. fructicola and C. siamense.
2.3. Culture and morphological characteristics
To observe macroscopic culture characteristics, the strains were cultured on PDA and incubated at 25 °C for one week. A mycelium plug (5 mm diameter) was taken from the actively growing edge of the colony and incubated on PDA at 25 °C for seven days under dark conditions. To observe microscopic characters, mounts of strains were made in 3% KOH (lactic acid). Measurements were performed using a light microscope (Nikon Eclipse 80i) for at least 30 conidia.
2.4. Pathogenicity tests
Pathogenicity tests were conducted to validate Koch’s postulates and establish a causative relationship between the fungal strain and disease. Fuji apple fruits were surface-sterilized using 1% sodium hypochlorite solution and were wounded with a 6 mm diameter cork borer. Each fruit was inoculated with mycelial plugs of each strain (6 mm diameter). Control fruits were treated with agar plugs (6 mm diameter) without fungal mycelium. The inoculated fruits were placed in a moistened container and incubated at 25 °C in dark. After 5 days, disease incidence was determined: no infection and symptom (−), 0.1–6 mm (+), 7–12 mm (++), and over 12 mm (+++). To fulfill Koch’s postulates, after seven days, each strain was re-isolated from the inoculated fruits following the above mention procedure, and re-identified using morphological features and multilocus sequence analysis.
3. Results
3.1. Phylogenetic analyses
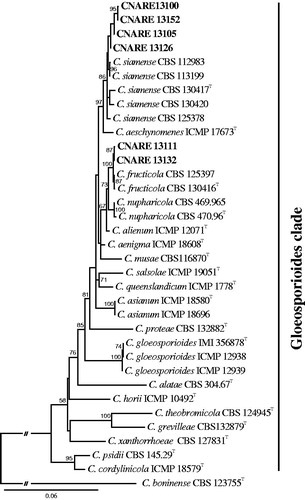
Based on the ITS sequence analysis and morphological features, six strains isolated from apples with bitter rot symptoms were identified as members of the C. gloeosporioides species complex (data not shown). Next, for the analysis of the concatenated dataset (ITS, ACT, CAL, CHS-1, and GAPDH) to identify the unknown strains to species level, we increased sampling of GenBank sequences of type strain in the C. gloeosporioides species complex. The corresponding lengths of the aligned fragments of six strains for the five loci were as follows: 582 bp for ITS; 275 bp for ACT; 708 bp for CAL; 286 bp for CHS-1; 283 bp for GAPDH. Two strains (CNARE13111 and CNARE13132) formed a monophyletic group with C. fructicola CBS 130416 (type strain from Coffea arabica) and CBS125397 (from Tetragastris panamensis) (100% bootstrap value), while four additional strains (CNARE13100, CNARE13105, CNARE13126, and CNARE13152) formed a monophyletic group with C. siamense CBS 130417 (type strain from Coffea arabica), CBS 112983 (Protea cynaroides), CBS 113199 (Protea cynaroides), CBS 125378 (Hymenocallis americana), CBS 130420 (Jasminum sambac) (86% bootstrap value) ().
Figure 1. Maximum likelihood phylogenetic tree based on the concatenated dataset (ITS, ACT, CAL, CHS-1, and GAPDH) used to identify Colletotrichum strains isolated from bitter rot on apple in Korea. Bootstrap scores greater than 50 are presented at the nodes. The scale bar indicates the number of nucleotide substitutions per site and the letter Tindicates ex-type strains. The strains originating from bitter rot on apple are indicated in bold.
3.2. Taxonomy
Colletotrichum fructicola Prihastuti, L. Cai & K.D. Hyde., Fungal Divers. 39: 89–109 (2009) ( ).
Figure 2. Colletotrichum fructicola CNARE13132 (a) and C. siamense CNARE13126 (b). (a) Colony morphology on potato dextrose agar (PDA) after 7-day culture at 25 °C (front); (b) Colony morphology on PDA after 7-day cultures at 25 °C (reverse); (c) Conidia Bar scale, 10 μm (micrometer); (d) Symptoms induced by artificial inoculation.
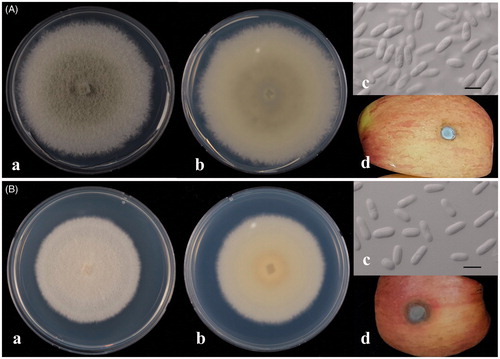
Colonies on PDA initially white mycelia that became grey to dark grey at the center with age on the front side and greyish green on the reverse side. Colonies grew 10.1–11.0 mm/day at 25 ± 1 °C under dark and were 70.7–77.0 mm diameter after seven days. Aerial mycelium dense, cottony pale grey, without visible conidial masses. Acervuli absent in culture. Setae absent. Conidia common in mycelium, one-celled, smooth-walled, hyaline, straight, aseptate, cylindrical with both ends rounded, sometimes oblong, sizes ranging 11.0–14.0 × 4.5–5.5 μm. Chlamydospores were not observed.
Remarks: Colletotrichum fructicola is morphologically similar to Colletotrichum kahawae, but it can be distinguished from C. kahawae by small conidia and rapid growth on PDA [Citation3].
Colletotrichum siamense Prihastuti, L. Cai & K.D. Hyde., Fungal Divers. 39: 89-109 (2009) ( ).
Colonies on PDA initially greyish white mycelia that became pale brownish to pinkish with age on the front side and pinkish on the reverse side. Colonies grew 9.3–10.3 mm/day at 25 ± 1 °C under dark and were 65.1–72.1 mm diameter after seven days. Aerial mycelium dense, cottony, greyish white, with visible conidial masses at inoculum. Setae absent. Conidia common in mycelium, one-celled, smooth-walled, hyaline, cylindrical with both ends bluntly rounded, sometimes oblong, sizes ranging 11.0–14.5 × 4.0–5.0 μm
Remarks: Colletotrichum siamense is morphologically similar to C. acutatum, but it can be distinguished from C. acutatum by cylindrical conidia with both ends bluntly rounded [Citation3].
3.3. Pathogenicity
Despite differences in virulence, all strains produced characteristic signs of bitter rot on apple fruit, such as a sunken zone with concentric rings of conidial masses. C. siamense (CNARE13100, CNARE13105, CNARE13126, and CNARE13152) produced larger lesions compared to C. fructicola (CNARE13111 and CNARE13132). Of the six strains isolated in this study, strain CNARE13126 of C. siamense showed the strongest virulence (; ).
4. Discussion
Apple fruits from Andong, Korea collected in 2013 showed characteristic signs of bitter rot – a sunken zone with concentric rings of conidial masses. Based on morphological features, multilocus sequence analyses, and pathogenicity tests, we identified C. fructicola and C. siamense to be the causative agents of bitter rot of apple fruit in Korea. To the best of our knowledge, this is the first report of C. fructicola and C. siamense in Korea causing bitter rot on apple fruit. C. fructicola and C. siamense, members of the C. gloeosporioides species complex, were first described as a pathogen of C. arabica berries [Citation15]. These species have wide host range including apple and are associated with bitter rot worldwide [Citation6].
Many new Colletotrichum species from various hosts were described using morphological features and multilocus phylogenetic analysis of ITS, ACT, CAL, CHS-1, and GAPDH [Citation1,Citation5,Citation6,Citation16]. C. gloeosporioides is known as one of the most important pathogens that infect a range plant species. However 25 isolates of C. gloeosporioides from tropical fruits were re-identified as C. asianum, C. fructicola, C. horii, C. kahawae, and C. gloeosporioides based on morphological characteristics and multilocus phylogenetic analysis of ITS, ACT, CAL, CHS-1, and GAPDH [Citation16]. Seven Collectotrichum species are known as causal species of bitter rot of apple (C. gloeosporioides, C. acutatum C. fioriniae, C. fructicola, C. nymphaeae, C. siamense, and C. theobromicola) [Citation1,Citation2]. C. gloeosporioides and C. acutatum were previously reported to cause bitter rot of apples in Korea (Korea disease list). These two species were not found in this study; we may have missed these species due to a low number of isolates in our study, or strains in previous studies may have been misidentified. Further study of bitter rot of apple fruit in Korea is needed to clarify this situation.
Bitter rot in Korea is caused by at least two Colletotrichum species, C. fructicola and C. siamense, with the latter being the more aggressive species. Studies of bitter rot and other plant pathogens should combine morphology multigene phylogenetic analysis for more reliable identification. We expect such an approach will uncover more fungal species associated with plant pathogens. In addition to more studies identifying causative agents of plant pathogens, studies of species distribution can provide valuable information for better management of bitter rot on apple in Korea.
Acknowledgments
We would like to thank Dr. Jonathan J. Fong (Lingnan University) for English editing and valuable advice.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Munir M, Amsden B, Dixon E, et al. Characterization of Colletotrichum species causing bitter rot of apple in Kentucky orchards. Plant Dis. 2016;100:2194–2203.
- González E, Sutton TB, Correll JC. Clarification of the etiology of Glomerella leaf spot and bitter rot of apple caused by Colletotrichum spp. based on morphology and genetic, molecular, and pathogenicity tests. Phytopathology 2006;96:982–992.
- Cai L, Hyde KD, Taylor PWJ, et al. A polyphasic approach for studying Colletotrichum. Fungal Divers. 2009;39:183–204.
- Jayawardena RS, Hyde KD, Damm U, et al. Notes on currently accepted species of Colletotrichum. Mycosphere. 2016;7:1192–1260.
- Damm U, Cannon PF, Woudenberg JHC, et al. The Colletotrichum acutatum species complex. Stud Mycol. 2012;73:37–113.
- Weir BS, Johnston PR, Damm U. The Colletotrichum gloeosporioides species complex. Stud Mycol. 2012;73:115–180.
- The Korean Society of Plant Pathology. List of plant diseases in Korea. 5th ed. Seoul: Korean Society of Plant Pathology; 2009.
- Uhm JY. Reduced fungicide spray program for major apple diseases Korea. Korea: Culture and Horticulture Press; 2010
- Lee DH, Kim D, Jeon Y, et al. Molecular and cultural characterization of Colletotrichum spp. causing bitter rot of apples in Korea. Plant Pathol J. 2007;23:37–44.
- Rogers SO, Bendich AJ. Extraction of total cellular DNA from plants, algae and fungi. In: Gelvin SB, Schilperoort RA, editors. Plant molecular biology manual. Dordrecht: Springer Netherlands; 1994. p. 183–190.
- Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739.
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780.
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014;30:1312–1313.
- Kornerup A, Wanscher JH. Methuen handbook of colour. London: Methuen; 1963.
- Prihastuti H, Cai L, Chen H, et al. Characterization of Colletotrichum species associated with coffee berries in northern Thailand. Fungal Divers. 2009;39:89–109.
- Phoulivong S, Cai L, Chen H, et al. Colletotrichum gloeosporioides is not a common pathogen on tropical fruits. Fungal Divers. 2010;44:33–43.
