Abstract
While evaluating fungal diversity in freshwater, grasshopper feces, and soil collected at Dokdo Island in Korea, four fungal strains designated CNUFC-DDS14-1, CNUFC-GHD05-1, CNUFC-DDS47-1, and CNUFC-NDR5-2 were isolated. Based on combination studies using phylogenies and morphological characteristics, the isolates were confirmed as Ascodesmis sphaerospora, Chaetomella raphigera, Gibellulopsis nigrescens, and Myrmecridium schulzeri, respectively. This is the first records of these four species from Korea.
1. Introduction
Fungi represent an integral part of the biomass of any natural environment including soils. In soils, they act as agents governing soil carbon cycling, plant nutrition, and pathology. Many fungal species also adapt to invade, colonize, and ensure decomposition of keratinous debris of other organisms [Citation1] and mineralization of herbivore feces [Citation2]. The distribution of the fungal community is related to different physiological, ecological, and bio-geographical features, closely linked to the surrounding terrestrial habitat [Citation3–5].
Dokdo Island is characterized by high soil salinity, drought, high winds, low amounts of organic matter, high concentrations of uric acid in the soil, and steep inclines [Citation6]. The weather usually features snow in the winter, with the climate influenced by warm ocean currents. Organisms living there have adapted to the environmental conditions by changing their genetic composition and enzymatic systems, and thus constitute a resource with great biotechnological potential. However, fungi occurring on this island remain poorly studied. Thus it is important to investigate the fungal diversity of this isolated area to understand their natural ecology. Reports have described a variety of bacterial and fungal species on Dokdo Island belonging to the genera Absidia, Alternaria, Aspergillus, Cladosporium, Clonostachys, Fusarium, Diaporthe, Metahizium, Mortierella, Mucor, Paecilomyces, Paraphoma, Penicillium, Plectosphaerella, and Stemphylium [Citation7–11]. However, comparatively few species of fungi have been described [Citation8–10].
Freshwater nourishes diverse habitats for fungi, such as fallen leaves, plant litter, decaying wood, aquatic plants and insects, and soils. Little information is available for fungi belonging to Basidiomycetes and Zygomycetes in comparison to freshwater Ascomycetes, which comprise approximately 622 species and 170 genera; more than 531 Hyphomycetes species (55 genera), and 183 species of Trichomycetes (3 orders) [Citation12]. Various studies related to fungal diversity from diverse habitats have been carried out by Korean mycologists. In comparison to the terrestrial environment, knowledge is scant in Korea regarding fungi belonging to freshwater habitats, particularly Ascomycetes. Freshwater fungi, especially Ascomycetes, are important in freshwater ecosystems as they provide nutrients for other aquatic microorganisms by decomposing complex organic compounds and because of enzymatic activity including that of cellulase, xylanase, and ligninase that degrades wood [Citation13]. Moreover, they are able to produce various compounds that act against pathogenic bacteria, fungi, and nematodes [Citation12].
Fungi in the fecal environment help to biodegrade organic materials and return the nutrients to the environment for reuse [Citation14]. Very little information has been published concerning the diversity of fungi in insect feces in comparison to that in various animal dung substrates [Citation15]. Melanoplus sanguinipes was isolated from grasshopper gut, but no detailed information was provided on the fungal communities [Citation16]. Among the fungal isolates from fecal samples in Korea, five species (two new species and three new records) were reported from rat feces and two (new records) from grasshopper. Absidia stercoraria, Mucor stercorarius, Absidia glauca, Paecilomyces variotti, and Cephaliophora tropica were reported from rat feces [Citation10,Citation17–20]. Cunninghamella echinulata and Albifimbria terrestris were reported from grasshopper feces [Citation19,Citation21]. Korean contributions to the occurrence of coprophilous fungi are still rare.
During an inventory of fungal species belonging to the classes Leotiomycetes, Pezizomycetes, and Sordariomycetes, four new records from freshwater, fecal, and soil samples were identified. The objective of the present study was to perform morphological and molecular analyses to characterize four undescribed species in Korea: Ascodesmis sphaerospora, Chaetomella raphigera, Gibellulopsis nigrescens, and Myrmecridium schulzeri.
2. Materials and methods
2.1. Isolation of fungal strains
Freshwater samples were collected from a branch stream of the Nakdong river located in Gyeongsangbuk-do, Korea. Grasshoppers were collected at the CNU Arboretum located in Chonnam National University, Gwangju, Korea. Soil samples were collected from Dokdo Island in July 2014. In this study, the isolation of fungi from freshwater, grasshoppers, and soil samples was performed as described previously [Citation10,Citation19]. Pure isolates were maintained in potato dextrose agar (PDA) slant tubes and stored in 20% glycerol at −80 °C at the Chonnam National University Fungal Collection (CNUFC), Gwangju, Korea. Strains CNUFC-DDS14-1, CNUFC-DDS47-1, and CNUFC-GHD05-1 were also deposited at the Culture Collection of the National Institute of Biological Resources (NIBR, Incheon, Korea), strain CNUFC-NDR5-2 was deposited at the Culture Collection of the Nakdonggang National Institute of Biological Resources (NNIBR, Sangju, Korea).
2.2. Morphological studies
To obtain samples for microscopic examination, CNUFC-DDS14-1, CNUFC-GHD05-1, CNUFC-DDS47-1, and CNUFC-NDR5-2 were cultured on PDA, corn meal agar (CMA: 20 g cornmeal, 20 g agar, 1 L distilled water), and malt extract agar (MEA: 20 g malt extract, 20 g agar, 1 L distilled water). Plates were incubated at 10, 20, 25, 30, and 35 °C in the dark for 7 days. Samples were mounted in lactophenol solution (Junsei Chemical Co. Ltd., Tokyo, Japan) and observed using a BX51 microscope equipped with DIC optics (Olympus, Tokyo, Japan).
2.3. DNA extraction, PCR, and sequencing
Genomic DNA was extracted directly from the mycelia of fungal isolates using the Solg TM Genomic DNA prep kit (Solgent Co. Ltd., Daejeon, South Korea). The rDNA ITS1-5.8S-ITS2 region, small subunit (SSU) 18S, and large subunit (LSU) 28S ribosomal DNA were amplified with the primer pairs ITS1/ITS4 [Citation22], NS1/NS4 [Citation23], and LROR/LR5F [Citation24]. The PCR amplification mixture (total volume, 20 μL) contained 10 ng of fungal DNA template, 5 pmol/μL of each primer, and Accupower PCR Premix (Taq DNA polymerase, dNTPs, buffer, and a tracking dye; Bioneer Corp., Daejeon, Korea). Purification of the PCR products was carried out using the Accuprep PCR Purification Kit (Bioneer Corp.) according to the manufacturer’s instructions. DNA sequencing was performed on an ABI 3700 Automated DNA sequencer (Applied Biosystems Inc., Foster City, CA).
2.4. Phylogenetic analysis
Fungal sequences obtained from the GenBank database () were aligned using Clustal_X v.1.83 [Citation25] and edited with Bioedit v.5.0.9.1 [Citation26]. Phylogenetic analyses were performed using MEGA 6 software [Citation27], and maximum likelihood (ML) was constructed by Kimura’s two-parameter correction method. The sequences of Acremonium alcalophilum, Caloscypha fulgens, Glomerella cingulata, Peziza succosa, Seiridium banksiae, and Smardaea amethystina were used as outgroups. The reliability of internal branches was assessed using the p-distance substitution model with 1000 bootstrap replications.
Table 1. Taxa, collection numbers, sequences, and GenBank accession numbers used in this study.
3. Results
3.1. Phylogenetic analysis
The results constructed by ML analyses of the four isolates are shown in . A Basic Local Alignment Search Tool (BLAST) search of ITS sequences via the NCBI database indicated that the isolates CNUFC-GHD05-1, CNUFC-DDS47-1, and CNUFC-NDR5-2 most closely resembled Chaetomella raphigera (GenBank accession no. AY487076), Gibellulopsis nigrescens (GenBank accession no. KP230818), and Myrmecridium schulzeri (GenBank accession no. KM056332) with 99.1% (332/335 bp), 99.8% (492/493 bp), and 100% (445/445 bp) homology, respectively. The 28S rDNA region of CNUFC-DDS14-1, CNUFC-GHD05-1, CNUFC-DDS47-1, and CNUFC-NDR5-2 revealed A. sphaerospora (GenBank accession no. FJ176858), C. raphigera (GenBank accession no. AY487077), G. nigrescens (GenBank accession no. GU180648), and M. schulzeri (GenBank accession no. EU041826) with similarities of 100% (372/372 bp), 99.6% (854/857 bp), 99.8% (849/851 bp), and 99.9% (795/796 bp), respectively. In addition, the BLASTn search results for the CNUFC-DDS14-1 18S rDNA showed 99.8% (1021/1031 bp) homology with A. sphaerospora (GenBank accession no. U53372).
Figure 1. Phylogenetic tree based on ML analysis of 18S and 28S sequences for Ascodesmis sphaerospora CNUFC-DDS14-1 and A. sphaerospora CNUFC-DDS14-2. The sequences of Caloscypha fulgens, Peziza succosa, and Smardaea amethystina were used as outgroups. Bootstrap support values of ≥50% are indicated at the nodes. The bar indicates the number of substitutions per position.
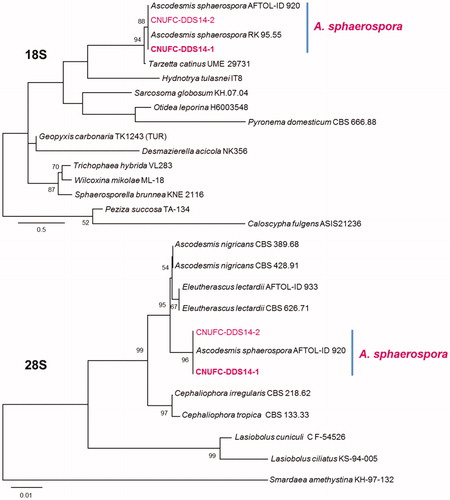
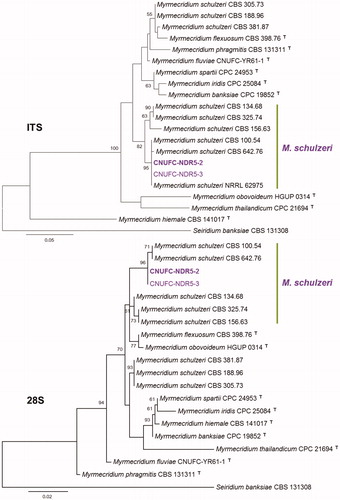
Figure 2. Phylogenetic tree based on ML analysis of internal transcribed rDNA and 28S sequences for Chaetomella raphigera CNUFC-GHD05-1 and C. raphigera CNUFC-GHD05-2. The sequence of Glomerella cingulata was used as an outgroup. Bootstrap support values of ≥50% are indicated at the nodes. The bar indicates the number of substitutions per position.
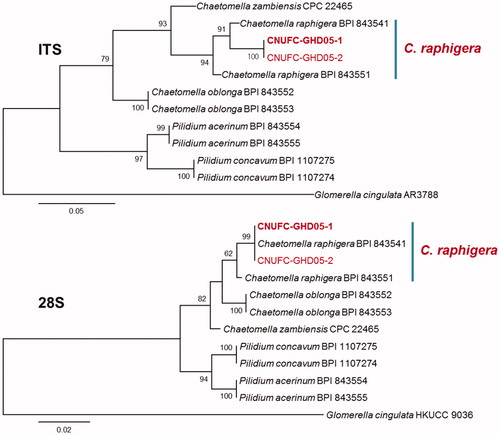
Figure 3. Phylogenetic tree based on ML analysis of internal transcribed rDNA and 28S sequences for Gibellulopsis nigrescens CNUFC-DDS47-1 and G. nigrescens CNUFC-DDS47-2. The sequence of Acremonium alcalophilum was used as an outgroup. Bootstrap support values of ≥50% are indicated at the nodes. The bar indicates the number of substitutions per position.
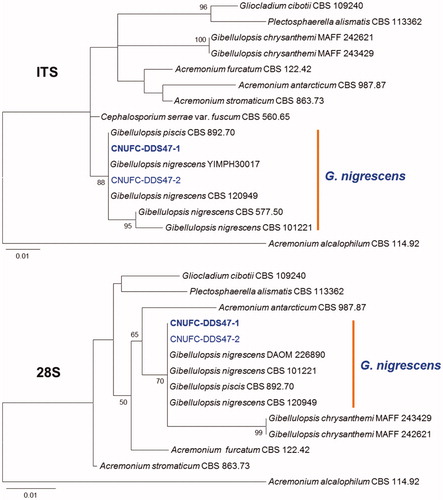
Figure 4. Phylogenetic tree based on ML analysis of internal transcribed rDNA and 28S sequences for Myrmecridium schulzeri CNUFC-NDR5-2 and M. schulzeri CNUFC-NDR5-3. The sequence of Seiridium banksiae was used as an outgroup. Bootstrap support values of ≥50% are indicated at the nodes. The bar indicates the number of substitutions per position.
3.2. Taxonomy
3.2.1. Taxonomy of CNUFC-DDS14-1
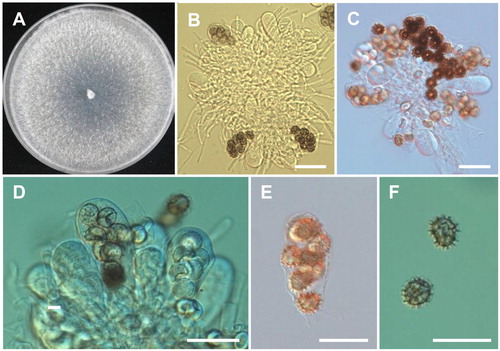
Ascodesmis sphaerospora W. Obrist, Canadian Journal of Botany 39:948 (1961) (; ).
Table 2. Morphological characteristics of CNUFC-DDS14-1 compared to those of the reference Ascodesmis sphaerospora strain.
Description: The strain grew rapidly at 25 °C on PDA, filling the petri dish after 4–5 days of incubation. The initial colony color was white and later turned to grayish white. Apothecia were superficial, sessile, and obconical. Asci were clavate, oblong, or ovoid, and measured 40–62 × 18–24 μm. Each asci contained 4–8 ascospores. Ascospores were one-celled, subglobose to ellipsoid, hyaline when young, becoming brown at maturity, and covered with dark brown markings in the form of spines, ridges, or reticulations, and measured 10–12 × 9–11 μm.
3.2.2. Taxonomy of CNUFC-GHD05-1
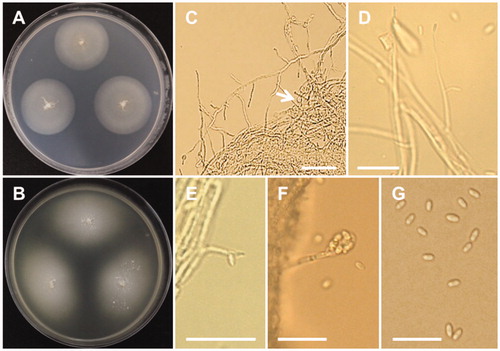
Chaetomella raphigera Swift, Mycologia 22:165 (1930) (; ).
Table 3. Morphological characteristics of CNUFC-GHD05-1 compared to those of the reference Chaetomella raphigera strain.
=Volutellospora raphigera (Swift) Thirum. & P.N. Mathur, Sydowia 18 (1-6):38 (1965).
=Chaetomella terricola P.Rama Rao, Mycopathologia et Mycologia Applicata 19 (3):255 (1963).
Description: Colonies of the strain grew slowly on PDA, reaching 20–22 mm in diameter at 25 °C after 7 days of incubation. The initial colony color was white and later turned to cinnamon. Pycnidia were elongated, reniform, pale to dark reddish brown, and measured 72.5–148.5 × 46.5–88.5 µm. Setae were pale to dark brown, mostly 2 septate, and measured 21.3–47.8 × 2.0–3.5 µm. Conidiophores were cylindrical, branched, 26.0–100 × 1.0–2.3 µm. Conidia were ellipsoid, and measured 4.8–7.2 × 1.8–2.6 µm.
3.2.3. Taxonomy of CNUFC-DDS47-1
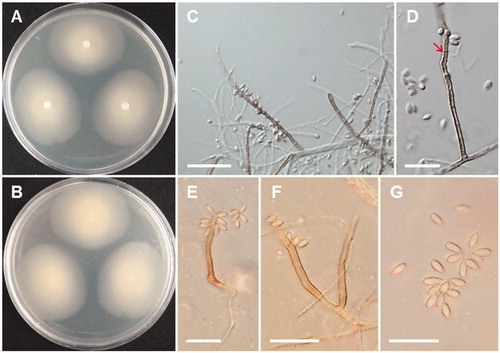
Gibellulopsis nigrescens R Zare, Gams W, Summerb, Nova Hedwigia 85:477 (2007) (; ).
Table 4. Morphological characteristics of CNUFC-DDS47-1 compared to those of the reference Gibellulopsis nigrescens strain.
≡Verticillium nigrescens Pethybr., Transactions of the British Mycological Society 6:117 (1919).
=Cephalosporium serrae Maffei, Atti dell'Istituto Botanico della Università e Laboratorio Crittogamico di Pavia 1:196 (1929).
=Verticillium amaranti Verona & Ceccar. (1935)
=Verticillium amaranthi Verona & Ceccar., Phytopathol. Z.:379 (1935).
=Verticillium dahliae f. zonatum J.F.H. Beyma, Antonie van Leeuwenhoek 6:42 (1940).
Description: Colonies of the strain grew slowly on PDA, reaching 25–27 mm in diameter at 20 °C after 10 days of incubation. The color of the colonies on PDA was whitish with cotton-like at the center. Conidiophores arise from vegetative hypha measuring 55–100 × 1.5–2.5 μm. Chlamydospores were formed as single or in short chains, and measured 4–6 × 2.5–5 μm. Conidia were smooth-walled and elongate-ellipsoidal, and measured 4.0–6.0 × 1.5–2.5 μm. Colony on MEA was light brown with 24–30 mm in diameter.
3.2.4. Taxonomy of CNUFC-NDR5-2
Myrmecridium schulzeri (Sacc.) Arzanlou, W. Gams & Crous, Studies in Mycology 58:84 (2007) (; ).
Table 5. Morphological characteristics of CNUFC-NDR5-2 compared to those of the reference Myrmecridium schulzeri strain.
≡Psilobotrys schulzerii Sacc. (1884).
≡Psilobotrys schulzeri Sacc., Hedwigia 23: 126 (1884).
≡Chloridium schulzeri (Sacc.) Sacc., Sylloge Fungorum 4: 322 (1886).
≡Rhinocladiella schulzeri (Sacc.) Matsush., Icones Microfungorum a Matsushima lectorum: 124 (1975).
≡Ramichloridium schulzeri (Sacc.) de Hoog, Studies in Mycology 15: 64 (1977).
=Acrotheca acuta Grove, Journal of Botany, British and Foreign 54: 222 (1916).
=Rhinotrichum multisporum Doguet, Revue Mycol., Paris: 78 (1952).
Description: Colonies of the strain grew slowly on MEA, reaching 29 mm diameter at 25 °C after 15 days of incubation. The colony color was pale orange. The colony reverse was also pale orange. Conidiophores were straight, almost unbranched, sometimes branched, reddish brown, septate, 2.5–3.5 μm in width, and variable in length. Conidiogenous cells were cylindrical and forming a rachis with scattered pimple-shaped denticles. Conidia were ellipsoid, obovoid, fusiform and measured 2.5–3.5 × 5.5–7.5 μm. On OA, the colonies grew more rapidly than on MEA and PDA, but abundant sporulation when grown on MEA.
4. Discussion
Here, we discussed morphological characteristics and the phylogeny of Ascodesmis sphaerospora, Chaetomella raphigera, Gibellulopsis nigrescens, and Myrmecridium schulzeri and compared these aspects to the most closely related species.
The genus Ascodesmis belonging to the class Pezizomycetes, order Pezizales, family Ascodesmidaceae, was first described by Van Tiegham [Citation28] with A. nigricans as the type species. This genus is of operculate discomycetes representing a primitive form of ascomycetes with no excipulum. According to Index Fungorum (www.indexfungorum.org), 13 species were assigned to this genus. There were no Ascodesmis species reported from Korea until this study. A. sphaerospora was first isolated from dung samples of Brazilian animals [Citation29]. This species is distinguished by its globose or subglobose ascospores with a reticulate ornamentation. A. sphaerospora shows a considerable variation in ascospores number, size, and shape growing on dung (spores are spherical) or on artificial agar media (spores tend to be more elliptical). These species are characterized by having relatively long spines [Citation29]. Comparing the morphology of the CNUFC-DDS14-1 isolate with previous descriptions by Obrist [Citation29], the present isolate was most similar to those of A. sphaerospora. In our molecular analyses, A. sphaerospora CNUFC-DDS14-1 formed a well-supported clade (). There is relatively little literature examining the molecular characteristics of Ascodesmis species in comparison to morphological identifications [Citation30,Citation31]. A. sphaerospora is reported to be isolated from the dung of jaguar, lion, ocelot, tiger, dog, elk, toad, rabbit, pig, and giraffe [Citation29,Citation32–34]. This is the first isolation of A. sphaerospora from a soil sample. A. sphaerospora produces antifungal and antibacterial metabolite arugosin F [Citation35].
The genus Chaetomella belonging to the class Leotiomycetes, order Helotiales, family Chaetomellaceae, was established by Fuckel in 1869, including C. atra and C. oblonga, based on the production of pycnidum fruiting body [Citation36]. The genus Chaetomella was designated considering its resemblance to genus Chaetomium, showing an external appearance of the fruiting bodies with characteristic appendages. Until now, 25 species of Chaetomella have been described according to Index Fungorum. Chaetomella spp. can be isolated from soil and plants [Citation37]. C. raphigera is reported to cause leaf spot disease to Cuphea spp., Rosa chinensis, blueberry, and pomegranate [Citation38–40]. Phylogenetic analysis based on ITS and LSU sequences showed that our strains CNUFC-GHD05-1 and CNUFC-GHD05-2 grouped together with C. raphigera (). In addition, C. raphigera CNUFC-GHD05-1 fits well with the description provided by Rossman et al. [Citation39]. C. raphigera was reported to produce pectinase, cellulase and xylanase activity, which could play a major virulence role for rot in pomegranates [Citation41]. Yoneda et al. [Citation42] reported that β-glucosidase secreted by C. raphigera. C. raphigera isolated from a medicinal plant Terminalia arjuna exhibit for potential production of natural anticancer drug taxol [Citation43]. This finding suggests that the strain CNUFC-GHD05-1 may be a useful source for biotechnological applications and should be investigated further.
The genus Gibellulopsis belonging to the class Sordariomycetes, order Glomerellales, family Plectosphaerellaceae, was established by Batista and Maia [Citation44], and was further reinvigorated by Zare et al. [Citation45]. It has hyaline, partly rather wide vegetative hyphae, verticillium-like conidiophores, and pigmented terminal or intercalary chlamydospores. These features distinguish it from the other related genera of the Plectosphaerellacae. Until now, three species (Gibellulopsis piscis, G. nigrescens, and G. chrysanthemi) have been described according to the Index Fungorum. The genus Gibellulopsis contains only one valid species, G. nigrescens [Citation46]. In 2012, G. chrysanthemi was isolated from a garland of Chrysanthemum leaves [Citation47]. G. nigrescens is a plant pathogen and can be isolated from soil and the lower parts of the stem of plants. The phylogeny formed by the separate ITS-rDNA and 28S-rDNA of CNUFC-DDS47-1 supports the taxonomic identification as G. nigrescens (). This species was also isolated from a soil sample and was shown to be the cause of wilt of sugar beets in China [Citation48,Citation49]. Compared with the morphological characters of G. nigrescens as described by Zare et al. [Citation45], the isolated strain CNUFC-DDS47-1 displays hyaline conidia that are smooth, ellipsoidal often in chains, and form abundant chlamydospores.
The genus Myrmecridium belonging to the class Sordariomycetes, order Amphisphaeriales, family Bartaliniaceae, was described by Arzanlou et al. [Citation50] with the type species being M. schulzeri. To date, 13 species of Myrmecridium have been described according to the Index Fungorum. The species belonging to this genus are characterized by the production of obovoid or fusiform conidia, tapering towards a narrowly truncate base, hyaline mycelium with pale to unpigmented, pimple-like denticles. They are frequently isolated from freshwater, soil, and plant tissue [Citation15,Citation51–53]. M. schulzeri SCSGAF0135 strain was reported to have antibacterial activity [Citation54]. In the phylogenetic tree (), CNUFC-NDR5-2 and CNUFC-NDR5-3 strains clustered with a strain putatively named M. schulzeri. Comparing the morphology of the CNUFC-NDR5-2 isolate with previous descriptions by Arzanlou et al. [Citation50], the present isolate was generally similar to those of M. schulzeri. However, some morphological features differed. The size of conidia described by Arzanlou et al. [Citation50] was larger [3–4 × (6–)9–10(–12) μm] than that (2.5–3.5 × 5.5–7.5 μm) observed in our isolate. Our M. schulzeri isolate presented conidiophores that were sometimes branched, which was not described by Arzanlou et al. [Citation50]. In a previous study, we found a new species, M. fluviae, from a freshwater sample in Korea [Citation20]. Our results suggest that freshwater habitats are a good source of Myrmecridium species.
This is the first report of Ascodesmis sphaerospora, Chaetomella raphigera, Gibellulopsis nigrescens, and Myrmecridium schulzeri in Korea. Future studies should investigate the ability of these species to produce extracellular enzymes as well as secondary metabolites.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Ajello L. Natural history of the dermatophytes and related fungi. Mycopathol Mycol Appl. 1974;53:93–110.
- Angel K, Wicklow DT. Relationships between coprophilous fungi and fecal substrates in a Colorado grassland. Mycologia. 1975;67:63–74.
- Kuthubutheen AJ, Webster J. Water availability and the coprophilous fungus succession. Trans Br Mycol Soc. 1986;86:63–76.
- Safar HM, Cooke RC. Explotation of faecal resource units by coprophilous Ascomycotina. Trans Br Mycol Soc. 1988;90:593–599.
- Safar HM, Cooke RC. Interactions between bacteria and coprophilous Ascomycotina and Coprinus species on agar and in copromes. Trans Br Mycol Soc. 1988;91:73–80.
- Ryu SH, Jang KH, Choi EH. Biodiversity of marine invertebrates on rocky shores of Dokdo. Korea Zool Stud. 2012;51:710–726.
- You YH, Yoon H, Kim H, et al. Plant growth-promoting activity and genetic diversity of endophytic fungi isolated from native plants in Dokdo Islands for restoration of a coastal ecosystem. Kor J Life Sci. 2013;23:95–101.
- Lee HW, Nguyen TT, Mun HY, et al. Confirmation of two undescribed fungal species from Dokdo of Korea based on current classification system using multi loci. Mycobiology. 2015;43:392–401.
- Ariyawansa HA, Hyde KD, Jayasiri SC, et al. Fungal Diversity Notes 111–252: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2015;75:27–274.
- Nguyen TT, Lee SH, Bae S, et al. Characterization of two new records of Zygomycete species belonging to undiscovered taxa in Korea. Mycobiology. 2016;44:29–37.
- You YH, Park JM, Seo YG, et al. Distribution, characterization, and diversity of the endophytic fungal communities on Korean seacoasts showing contrasting geographic conditions. Mycobiology. 2017;45:150–159.
- Jones EBG, Hyde KD, Pang KL. Freshwater Fungi and fungal-like organisms. Marine and Freshwater Botany. Berlin-Boston: Walter de Gruyter; 2014.
- Bucher VVC, Pointing SB, Hyde KD, et al. Production of wood decay enzymes, loss of mass, and lignin solubilization in wood by diverse tropical freshwater fungi. Microb Ecol. 2004;48:331–377.
- Angel K, Wicklow DT. Decomposition of rabbit faeces: an indication of the significance of the coprophilous microflora in energy flow schemes. J Ecol. 1974;62:429–437.
- Thilagam L, Nayak BK, Nanda A. Studies on the diversity of coprophilous microfungi from hybrid cow dung samples. Int J Pharm Tech Res. 2015;8:135–138.
- Mead LJ, Khachatourians GG, Jones GA. Microbial ecology of the gut in laboratory stocks of the migratory grasshopper, Melanoplus sanguinipes (Fab.) (Orthoptera: Acrididae). Appl Environ Microbiol. 1988;54:1174–1181.
- Li GJ, Hyde KD, Zhao RL, et al. Fungal diversity notes 253-366: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016;78:1–237.
- Nguyen TT, Paul NC, Lee HB. Characterization of Paecilomyces variotii and Talaromyces amestolkiae in Korea based on the morphological characteristics and multigene phylogenetic analyses. Mycobiology. 2016;44:248–259.
- Nguyen TTT, Choi Y-J, Lee HB. Three unrecorded fungal species from fecal and freshwater samples in Korea. Kor J Mycol. 2017;45:304–318.
- Tibpromma S, Hyde KD, Jeewon R, et al. Fungal diversity notes 491-602: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2017;83:1–261.
- Nguyen TT, Choi YJ, Lee HB. Isolation and characterization of three unrecorded Zygomycete fungi in Korea: Cunninghamella bertholletiae, Cunninghamella echinulata, and Cunninghamella elegans. Mycobiology. 2017;45:318–326.
- White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York, NY: Academic Press, Inc.; 1990. p. 315–322.
- Lee J, Lee S, Young JPW. Improved PCR primers for the detection and identification of arbuscular mycorrhizal fungi. FEMS Microbiol Ecol. 2008;65:339–349.
- Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172:4238–4246.
- Thompson JD, Gibson TJ, Plewniak F, et al. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882.
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98.
- Tamura K, Stecher G, Peterson D, et al. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729.
- Van Tieghem P. Sur le développement du fruit des Ascodesmis, genre nouveau de l'ordre des Ascomycètes. Bull Soc Bot France. 1876;23:271–279.
- Obrist W. The genus Ascodesmis. Can J Bot. 1961;39:943–945.
- Landvik S, Egger KN, Schumacher T. Towards a subordinal of the Pezizales (Ascomycota): phylogenetic analyses of SSU rDNA sequences. Nord J Bot. 1997;17:403–418.
- Hansen K, Perry BA, Pfister DH. Phylogenetic origins of two cleistothecial fungi, Orbicula parietina and Lasiobolidium orbiculoides, within the operculate discomycetes. Mycologia. 2005;97:1023–1033.
- Van Brummelen J. The genus Ascodesmis (Pezizales, Ascomycetes). Persoonia. 1981;11:333–358.
- Kristiansen R. The genus Ascodesmis (Pezizales) in Norway. Ascomycete. Org. 2011;2:65–69.
- Jeamjitt O, Manoch L, Visarathanonth N, et al. Coprophilous ascomycetes in Thailand. Mycotaxon. 2007;100:115–136.
- Hein SM, Gloer JB, Koster B, et al. Arugosin F: a new antifungal metabolite from the coprophilous fungus Ascodesmis sphaerospora. J Nat Prod. 1998; 61:1566–1567.
- Fuckel L. Symboiae Mycologicae. Nassau Ver Naturk. 1869;402:23–24.
- Stolk AC. The genus Chaetomella Fuckel. Trans Br Mycol Soc. 1963;46:409–425.
- Singh HB, Johri JK, Singh M, et al. A new leaf spot disease of Cuphea spp. caused by Chaetomella raphigera. Bull OEPP. 1999;29:213–214.
- Rossman AY, Aime MC, Farr DF, et al. The coelomycetous genera Chaetomella and Pilidium represent a newly discovered lineage of inoperculate discomycetes. Mycol Progress. 2004;3:275–290.
- Zhang M, Li JJ, Wu HY, et al. First report of Chaetomella raphigera causing leaf spot on Rosa chinensis in China. Plant Dis. 2014;98:569.
- Gajbhiye M, Sathe S, Shinde V, et al. Morphological and molecular characterization of pomegranate fruit rot pathogen, Chaetomella raphigera, and its virulence factors. Indian J Microbiol. 2016;56:99–102.
- Yoneda A, Kuo HWD, Ishihara M, et al. Glycosylation variants of a β-glucosidase secreted by a Taiwanese fungus, Chaetomella raphigera, exhibit variant specific catalytic and biochemical properties. PloS ONE. 2014;9:e106306. DOI:10.1371/journal.pone.0106306
- Gangadevi V, Muthumary J. A novel endophytic Taxol-producing fungus Chaetomella raphigera isolated from a medicinal plant, Terminalia arjuna. Appl Biochem Biotechnol. 2009;158:675–684.
- Batista AC, Maia HDS. Uma nova doença fungica de peixe ornamental. Anais Soc Biol Pernambuco. 1959;16:153–159.
- Zare R, Gams W, Starink-Willemse M, et al. Gibellulopsis, a suitable genus for Verticillium nigrescens and Musicillium, a new genus for V. theobromae. Nova Hedw. 2007;85:463–489.
- Seifert K, Morgan-Jones G, Gams W, et al. The genera of hyphomycetes. CBS Biodiversity Series. Utrecht (Netherlands): CBS–KNAW Fungal Biodiversity Centre; 2011. p. 997.
- Hirooka Y, Kawaradani M, Sato T. Description of Gibellulopsis chrysanthemi sp. nov. from leaves of garland chrysanthemum. Mycol Progress. 2014;13:13–19.
- Wu YM, Xu JJ, Wang HF, et al. Geosmithia tibetensis sp. nov. and new Gibellulopsis and Scopulariopsis records from Qinghai-Tibet. Mycotaxon. 2013;125:59–64.
- Zhou Y, Zhao ZQ, Guo QY, et al. First report of wilt of sugar beet caused by Gibellulopsis nigrescens in the Xinjiang region of China. Plant Dis. 2017;101:1318.
- Arzanlou M, Groenewald JZ, Gams W, et al. Phylogenetic and morphotaxonomic revision of Ramichloridium and allied genera. Stud Mycol. 2007;58:57–93.
- Crous PW, Summerell BA, Shivas RG, et al. Fungal planet description sheets: 92-106. Persoonia. 2011;27:130–162.
- Jiea CY, Zhoua QX, Zhao WS, et al. A new Myrmecridium species from Guizhou, China. Mycotaxon. 2013;124:1–8.
- Peintner U, Knapp M, Fleischer V, et al. Myrmecridium hiemale sp. nov. from snow-covered alpine soil is the first eurypsychrophile in this genus of anamorphic fungi. Int J Syst Evol Microbiol. 2016;66:2592–2598.
- Zhang XY, Bao J, Wang GH, et al. Diversity and antimicrobial activity of culturable fungi isolated from six species of the South China Sea gorgonians. Microb Ecol. 2012;64:617–627.