Abstract
Agrobacterium tumefaciens-mediated transformation (ATMT), as a simple and versatile method, achieves successful transformation in the yeast-like cells (YLCs) of Tremella fuciformis with lower efficiency. Establishment of a more efficient transformation system of YLCs is important for functional genomics research and biotechnological application. In this study, an enzymolysis-assisted ATMT method was developed. The degradation degree of YLCs depends on the concentration and digestion time of Lywallzyme. Lower concentration (≤0.1%) of Lywallzyme was capable of formation of limited wounds on the surface of YLCs and has less influence on their growth. In addition, there is no significant difference of YLCs growth among groups treated with 0.1% Lywallzyme for different time. The binary vector pGEH under the control of T. fuciformis glyceraldehyde-3-phosphate dehydrogenase gene (gpd) promoter was utilized to transform the enzymolytic wounded YLCs with different concentrations and digestion time. The results of PCR, Southern blot, quantitative real-time PCR (qRT-PCR) and fluorescence microscopy revealed that the T-DNA was integrated into the YLCs genome, suggesting an efficient enzymolysis-assisted ATMT method of YLCs was established. The highest transformation frequency reached 1200 transformants per 106 YLCs by 0.05% (w/v) Lywallzyme digestion for 15 min, and the transformants were genetically stable. Compared with the mechanical wounding methods, enzymolytic wounding is thought to be a tender, safer and more effective method.
1. Introduction
Tremella fuciformis is a famous culinary-medicinal jelly mushroom cultivated in China and other Southeast Asia countries [Citation1,Citation2]. The yeast-like cells (YLCs) of T. fuciformis are asexually reproduced from basidiospores by budding. The YLCs are ideal material for genetic manipulation and gene functional research, they could also be used as a microbial cell factory system [Citation3–7]. To this end, an efficient genetic transformation protocol for YLCs is increasingly needed.
In previous attempts, several transformation methods have been developed for the YLCs including electroporation [Citation4,Citation8,Citation9], restriction enzyme mediated integration (REMI) [Citation3], polyethylene glycol (PEG)-mediated protoplast transformation (PMT) [Citation6,Citation10] and Agrobacterium tumefaciens-mediated transformation (ATMT) [Citation5,Citation7,Citation11–13]. Among these methods, ATMT is a relatively simple and effective genetic transformation system. However, the transformation efficiency is still lower. A possible reason is that the intact YLCs have a thick cell wall consisting of layers of chitin, β-glucan, and glycoproteins [Citation14], which increases the difficulty to absorb the T-DNA.
It was reported that the efficiency of ATMT was improved by wounding the receptor cells. Mechanical wounding of mycelia by vortexing with the existence of aluminium oxide particles obtained relatively higher transformation efficiency in several medicinal mushroom species, such as Ganoderma lucidum, Lentinula edodes, Grifola frondosa, Schizophyllum commune, and Lyophyllum decastes [Citation15]. Transformation efficiency was also improved by wounding the YLCs of T. fuciformis via vortexing [Citation12]. In addition, chemical wounding by NaOH treatment was able to generate cell surface wounds in T. fuciformis, which made them susceptible to ATMT [Citation13].
Cell wall lyticases have been successfully applied to protoplast preparation in a variety of fungi [Citation16]. The degradation degree of fungal cell wall depends mainly on the concentration and digestion time of the lyticase selected. Limited wounds of cell walls by enzymolysis might have less influence on cells’ growth but facilitate T-DNA transfer, which would improve the transformation efficiency. Here we investigated a suitable degradation condition for limited wounds to establish a relatively efficient enzymolysis-assisted ATMT method for the YLCs of T. fuciformis.
2. Materials and methods
2.1. Strains, plasmid, and growth conditions
T. fuciformis isolate Y32 (YLCs), preserved in the laboratory of Food Microbiology, Huazhong Agricultural University, was maintained on potato dextrose agar (PDA) and sub-cultured into the complete medium (CM) (20 g glucose, 1.32 g (NH4)2SO4, 0.25 g MgSO47H2O, 0.5 g KH2PO43H2O, 0.2 mg vitamin B1, 2 mg ZnSO47H2O, 0.5 g CaCl22H2O, 0.02 mg ammonium molybdate per litter) at 25 °C on an orbital shaker (150 rpm) (Max Q800; Thermo Fisher Scientific, Waltham, MA) in the dark. A binary plasmid pGEH, carring the enhanced green fluorescent protein gene (egfp)-hygromycin B phosphotransferase gene (hph) fusion expression cassette under the drive of the native glyceraldehyde-3-phosphate dehydrogenase gene (gpd) promoter, was constructed in our previous study [Citation7]. A. tumefaciens strain EHA105 haboring pGEH, grown in YEB medium (5 g tryptone, 1 g yeast extract, 5 g nutrient broth, 5 g sucrose, 0.49 g MgSO47H2O per litter) containing kanamycin (50 μg/ml) and rifampicin (50 μg/ml), was used to transform the YLCs. The transformed colonies were maintained on the potato dextrose selected agar (PDSA) under selection pressure (50 μg/ml hygromycin B and 200 μg/ml cefotaxime).
2.2. Influence of lyticase to YLCs growth
Lywallzyme (Guangdong Institute of Microbiology, Guangdong, China), an effective complex cell wall lyticase used for edible fungal protoplast isolation [Citation17,Citation18], was resolved in double distilled water and sterilized by a 0.22-μm bacterial filter membrane. Three-day-old YLCs were diluted by double distilled water to 106 cells/ml. Lywallzyme concentration and digestion time were chosen to evaluate the influence of Lywallzyme to YLCs growth by using single factor test. Equal volume of YLCs suspension and different concentrations of Lywallzyme solution (0, 0.0125%, 0.025%, 0.05%, 0.1%, 0.2%, 0.4%, 0.6%, 0.8%, 1.6%) were mixed and then digested in a 32 °C water bath for 15 min. Subsequently, fixed the Lywallzyme concentration, YLCs were digested at 32°Cfor different time (0, 15 min, 30 min, 45 min, 60 min, 75 min, 90 min, 105 min, 120 min). The survived YLCs was enumerated on PDA plates after 7 days’ cultivation at 25 °C.
2.3. Observation of wounded YLCs by scanning electron microscope
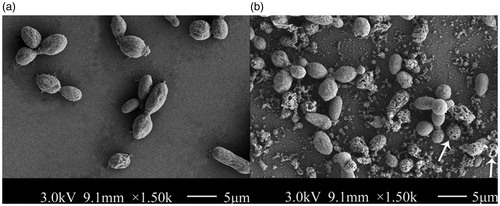
According to the aforementioned influence result of lyticase to YLCs growth, three-day-old YLCs were treated with the maximal effective concentration of Lywallzyme and then collected by centrifugation at 8000 rpm for 5 min and washed with PBS three times. After treatment with gradient dehydration and critical point drying, the YLCs were observed under a field emission scanning electron microscope (SEM) (Model SU8010; Hitachi, Tokyo, Japan).
2.4. Enzymolysis-assisted Agrobacterium tumefaciens-mediated transformation
Lywallzyme concentration and digestion time were chosen to evaluate the influence of enzymolysis wounds to transformation efficiency using single factor test. The manipulation steps were the same as the methods described in 2.3. YLCs (3 days) were diluted into 106 cells/ml by IM. Equal volume of YLCs suspension and different concentrations of Lywallzyme solution (0, 0.0125%, 0.025%, 0.05%, 0.1%, 0.2%, 0.4%) were mixed and then digested in a 32 °C water bath for 15 min. Subsequently, fixed the Lywallzyme concentration, YLCs were digested at 32 °C for different time (0, 15 min, 45 min, 90 min, 120 min).
A. tumefaciens strain EHA105 haboring pGEH was cultivated at 28 °C in YEB medium containing kanamycin (50 μg/ml) and rifampicin (50 μg/ml) to an OD600 of 0.6–0.8. Then bacterial cells were resuspended in induced medium (IM) with 200 μM acetosyringone (AS) (Sigma-Aldrich, St. Louis, MO) to an OD600 of 0.15–0.2 and incubated for another 6 h at 28 °C on an orbital shaker (180 rpm) for virulence pre-induction. 500 μl of wounded YLCs suspension were mixed with 500 μl of virulence pre-induced A. tumefaciens suspension and spread onto the mixed cellulose esters membrane (MCEM) on solid IM (with 200 μM AS) at 28 °C for 3 days. After incubation, the membranes with fungal and bacterial colonies were transferred onto PDSA, which was added with 200 μg/ml cefotaxime to kill the A. tumefaciens and 50 μg/ml hygromycin B to select the putative transformants. After 15 d, the colonies grew on the membrane were identified as putative transformants.
2.5. Verification of transformants
2.5.1. DNA isolation and PCR
Genomic DNA was extracted from five-round subcultured putative transformants by using the cetyltrimethyl ammonium bromide (CTAB) and a phenol-chloroform DNA extraction protocol omitting the β-mercaptoethanol [Citation19]. To confirm the existence of hph gene and egfp gene in transformants, PCR analysis was performed by using the following primer pairs: hph-F and hph-R; egfp-F and egfp-R, respectively (). The amplification procedures were carried out as follows: an initial denaturation at 94 °C for 3 min; 34 cycles of 94 °C denaturation for 30 s, 55 °C annealing for 30 s, 72 °C elongation for 1 min; and a final extension at 72 °C for 10 min. PCR products were analyzed by 1.0% agarose gel electrophoresis.
Table 1. Primers for PCR amplification in this work.
2.5.2. Southern blotting
To analyze the number of T-DNA copies integrated into the YLCs genome, putative transformants were chosen randomly to perform the Southern blot analysis. Wild-type Y32 was used as the negative control and the pGEH extracted by using the E.Z.N.ATM Plasmid mini Kit I (OMEGA Bio-tek, Norcross, GA) was used as the positive control. The hph fragment labeled with DIG was used as a specific probe for detecting. Total DNA (approximately 10 μg per sample) was digested with XhoI (Takara, Shiga, Japan) for 12 h at 37 °C. Then the product was separated on a 1% agarose gel (Biowest, Barcelona, Spain) and transferred onto the nylon membranes. Hybridization and detection were conducted according to the instructions of the DIG-High Prime DNA Labeling and Detection Kit (Roche, Mannheim, Germany).
2.6. Expression of reporter gene in transformants
2.6.1. Quantitative real-time PCR analysis
To test the relatively expression ratio of the egfp gene, total RNA was extracted from T-DNA integrated transformants by means of RNaiso plus (Takara) according to the manufacturer’s protocol. The first strand of cDNA was prepared with 5 μg of total RNA using the M-MLV RTase cDNA synthesis kit (Takara). Quantitative real-time PCR (qRT-PCR) was performed to test the relatively expression ratio of the egfp gene with the ABI ViiA7 Real-Time PCR System (Applied Biosystems, Foster City, CA). β-tubulin gene was as the endogenous control and the qRT-PCR conditions were 95 °C for 1 min, followed by 40 cycles of 95 °C for 30 s, and 60 °C for 30 s. The primer sequences for egfp gene and β-tubulin gene are listed in . Each reaction was replicated three times to estimate error and the gene expression was normalized by the 2−ΔΔCt analysis.
2.6.2. Fluorescence microscopy
For eGFP detection, different egfp expression level transformants were selected and mounted in double distilled water and then were analyzed by using a Nikon Eclipse 80i fluorescence microscope (Nikon, Tokyo, Japan) with a B-2A filter. The samples were excited at 460–500 nm wavelengths at automatic exposure time. Photomicrographs were captured under 40x objective for random transformants and processed with NIS-Elements BR 3.0 imaging software (Nikon). Wide-type Y32 was used as a negative control.
2.7. Detection of the stability of transformants
To determine whether the inserted T-DNA was integrated into the transformants genome stably, the transformants were cultured on PDA for five successive generations. Thereafter, the transformants were transferred to PDA containing 50 μg/ml hygromycin B to observe whether the colonies survived under the antibiotic pressure again.
3. Results
3.1. Enzymolytic wounding conditions for YLCs
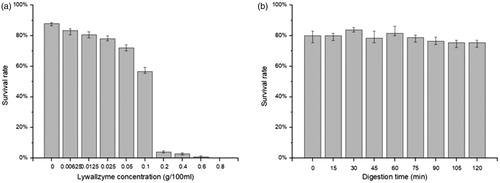
The cell growth was influenced by higher concentration Lywallzyme treatment seriously. The majority of YLCs cannot grow on the PDA with above 0.1% Lywallzyme treatment (). In addition, the cell survival rate did not show significant differences when the YLCs were treated with 0.1% Lywallzyme for different digest time (). The degradation degree of YLC wall depends on the concentration and digestion time of Lywallzyme. No wounds can be observed on the surface of YLCs without Lywallzyme treatment (). The YLCs were elliptic or fusiform observed under a SEM and the size of these cells were approximately 9 μm in diameter (). However, lower concentration (≤0.1%) Lywallzyme treatment led to the formation of limited wounds on the surface of YLCs and had little influence on YLCs growth ().
3.2. Integration of T-DNA in transformants
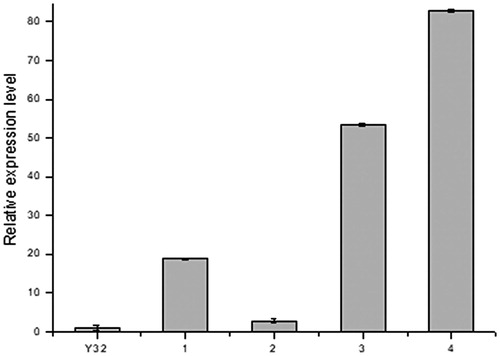
PCR analysis showed that the 7 transformants yielded the expected 717 bp and 1026 bp target bands (). From the results of Southern blot, we found that no hybridizing fragment was produced on the membrane in blank group, Y32 wild type. In 8 randomly selected transformants, most of transformants yielded single band of different sites, indicating random and single-copy insertion of T-DNA into the fungal genome. Meanwhile, only one double-copy transformant and one triple-copy transformant were obtained (). The results of quantitative real-time PCR (qRT-PCR) indicated that egfp gene was expressed with different levels and the copy number is not proportional to the expression level ().
Figure 3. PCR electrophorogram of YLCs of T. fuciformis transformants. (a) egfp fragments amplified by PCR. (b) hph fragments amplified by PCR M: Trans2K plus # DNA marker (Transgen, Beijing, China); −: wild-type T. fuciformis strain Y32 (negative control), +: vector pGEH (positive control); 1–8: randomly selected transformants.
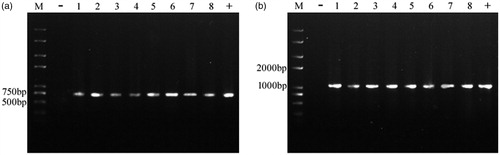
Figure 4. Southern blot analysis of transformants of T. fuciformis YLCs. Genomic DNA isolated from hygromycin-resistant transformants (10 μg) was digested with XhoIand probed with a fragment of the hph gene. M: λ-Hind III digest DNA marker (Takara, Japan); −: wild-type T. fuciformis strain Y32 (negative control); +: vector pGEH (positive control); 1–8: hygromycin-resistant transformants.
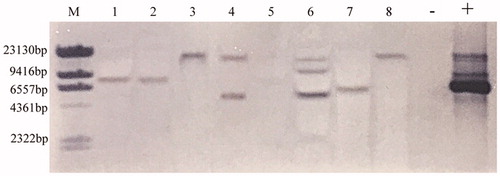
Figure 5. Relative expression ratio of the egfp in transformants of T. fuciformis YLCs and wild-type strain Y32. 1, 2: single-copy transformant; 3: double-copy transformant; 4: triple-copy transformant. Data were presented as mean values of three replicates with the corresponding standard deviations.
3.3. Transformation efficiency of the enzymolysis-assisted ATMT
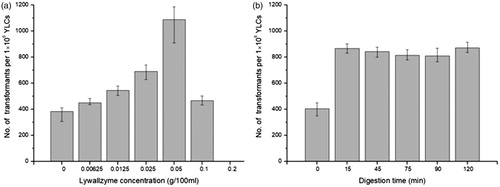
Transformation efficiency increased with the increase of the enzyme concentration (≤0.05%). The highest efficiency, 1200 transformants per 106 YLCs was obtained when the Lywallzyme concentration is up to 0.05%, which is 3-fold than that of the control group. However, the efficiency declined sharply when higher concentration of Lywallzyme (≥0.1%) was used (), no significant differences were found for different treatment time ().
3.4. Expression of eGFP in transformants
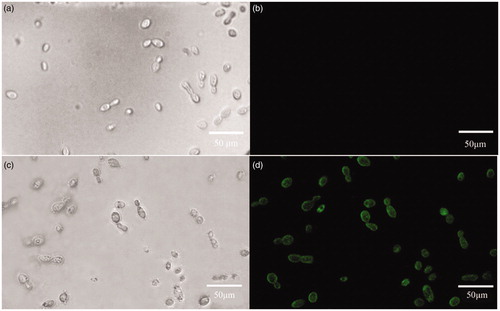
Green fluorescence was not found in wild-type strain, but observed in individual transformed colonies under the dark-field microscope (). Observing strong green fluorescence revealed that transformation was achieved successfully.
Figure 7. Expression of eGFP in individual transformants of T. fuciformis YLCs with hygromycin. (a) Wild type T. fuciformis strain Y32 (bright field). (b) Wild type T. fuciformis strain Y32 (corresponding fluorescence field). (c) Transformant (bright field). (d) Transformant (corresponding fluorescence field). Detection for bright field micrographs and corresponding fluorescence under UV light are shown with the same microscope equipment of excitation filters at 450–490 nm, a dichroic filter at 505 nm and an emission filter at 520 nm. Images were taken with 40x fields of view, bar = 50 μm.
3.5. Stability of transformants
8 transformants selected randomly were transferred onto the non-selection PDA medium and then back onto the selection one, they were still able to grow and their colony morphologies were no discernible difference. It indicated that these transformants preserved the hyh gene and were mitotically stable.
4. Discussion
Numerous efforts have been made to increase the transformation efficiency of YLCs in T. fuciformis. Mechanical wounding-assisted ATMT method obtained near 350 transformants per 106 YLCs [Citation12]. NaOH treatment ATMT method produced 40–50 transformants per 106 YLCs in average [Citation13]. However, the transformation efficiency by enzymolytic wounding reached more than 1200 transformants per 106 YLCs in current research. It is approximately 3-fold higher than that of control group using intact YLCs. The results demonstrated that Lywallzyme treatment is effective to increase the transformation frequency. A reasonable interpretation is that the wounds digested by Lywallzyme increase the possibility of the T-DNA intake. Lywallzyme was usually used for protoplast preparation via dissolving the fungal cell walls. In this study, Lywallzyme was resolved in double distilled water to avoid the formation of protoplast survived in iso-osmotic solution. Lower concentration and shorter time treatment of Lywallzyme lead to limited wounds of YLCs, which was not enough to produce protoplasts and had less influence on the growth of YLCs. The limited wounds on the surface of YLCs make them susceptible to ATMT after Lywallzyme treatment.
With the rise of the Lywallzyme concentration, the survival rate of the YLCs deceased. It was attributed to the accumulation of wounds in YLCs. However, less than two-hour incubation of 0.1% Lywallzyme, the survival rate showed no significant decrease with the prolonged incubation.
Different promoters on the binary vectors also have impact on the transformation efficiency. CaMV35S promoter, widely used in plant transformation, is a heterogenous promoter for T. fuciformis. No transformants were obtained via pCAMBIA 1300 carrying hph gene under the drive of CaMV35S in YLCs transformation [Citation12]. gpd promoter is an enhanced constitutive promoter [Citation20]. In our study, a native gpd promoter from T. fuciformis was used to carry out successful transformation. It was reported that homologous gpd promoter region is effective in directing the heterologous genes expression among several mushrooms. The Lentinus edodes gpd promoter and hph gene were fused together to allow a strong expression of a heterologous gene in L. edodes [Citation21]. gpd promoter isolated from Flammulina velutipes was used to construct vector for high-level heterologous genes expression [Citation22]. The hph gene under control of the promoter gpd-Le from L. edodes was more effectively integrated into the genome in T. fuciformis than that with the promoter gpd-An from Aspergillus nidulans [Citation10]. It revealed that suitable match between promoters and host cells was beneficial for effective expression of foreign genes in host cells. Homologous promoter or promoter from organisms close in genetic relationship is an efficient cis-element that can be easily recognized by regulatory factors of recipient hosts, resulting in methylation reduction, integration promotion and foreign gene expression.
5. Conclusion
In this study, an efficient enzymolysis-assisted A. tumefaciens-mediated transformation method for the YLCs of T. fuciformis was established. The hph and egfp gene were successfully introduced into the genomes of the YLCs. The transformation efficiency reached 1200 transformants per 106 YLCs with 0.05% (w/v) Lywallzyme digestion for 15 min, and the transformants were genetically stable. Compared with the mechanical wounding methods, enzymolytic wounding is thought to be a tender, safer, and more effective method.
Disclosure statement
The authors declare no conflict of interest related to any part of the research.
Additional information
Funding
References
- Cheung PCK. The hypocholesterolemic effect of two edible mushrooms: Auricularia auricula (tree-ear) and Tremella fuciformis (white jelly-leaf) in hypercholesterolemic rats. Nutr Res. 1996;16:1721–1725.
- Gao Q, Seljelid R, Chen H, et al. Characterisation of acidic heteroglycans from Tremella fuciformis berk with cytokine stimulating activity. Carbohydr Res. 1996;288:135–142.
- Zhu H, Wang TW, Sun SJ, et al. Chromosomal integration of the Vitreoscilla hemoglobin gene and its physiological actions in Tremella fuciformis. Appl Microbiol Biotechnol. 2006;72:770–776.
- Sun SJ, Chen DX, Xie BG, et al. Isolation of gpd promoter from Tremella fuciformis and driving expression of egfp gene. DNA Cell Biol. 2009;28:65–70.
- Shin DI, Song KS, Park HS. Oral vaccination of mice with Tremella fuciformis, yeast-like conidium cells expressing HBsAg. Biotechnol Lett. 2015;37:539–544.
- Kang L, Li Q, Lin J, et al. Biosynthesis of resveratrol in blastospore of the macrofungus Tremella fuciformis. Mol Biotechnol. 2015;57:675–684.
- Zhu H, Liu D, Wang Y, et al. Use of the yeast-like cells of Tremella fuciformis as a cell factory to produce a Pleurotus ostreatus hydrophobin. Biotechnol Lett. 2017;39:1167–1173.
- Guo L, Liu Y, Zhao S, et al. Highly efficient transformation of intact yeast-like conidium cells of Tremella fuciformis by electroporation. Sci China Ser C. 2008;51:932–940.
- Zhu J, Rao Y, Xie B, et al. Transformation of human lactoferrin gene to Tremella fuciformis. Chin J Tropical Crops (Chinese). 2010;31:1137–1142.
- Guo L, Liu E, Wang J, et al. Development of highly efficient transformation system of yeast-like conidia of Tremella fuciformis. Agr Sci China. 2009;8:268–275.
- Dong J, Ren D, Zheng L, et al. Establishment of efficient transformation system via Agrobacteriun tumefaciens in Tremella fuciformis. In: Proceedings of Asian Mycological Congress 2013 and the 13th International Marine and Freshwater Mycology Symposium Abstract Book; 2013 Aug 19-23; Beijing, China. Beijing, (China): China Academic Journal Electronic Publishing House; 2017. p. 168–169.
- Shin DI, Park HS. Mechanical wounding of yeast-like conidium cells of makes them susceptible to Agrobacterium-mediated transformation. Biosci Biotech Bioch. 2013;77:2157–2159.
- Shin DI, Park HS. Genetic transformation of the mycelia of Tremella fuciformis and changes of cytotoxicity. Kor J Mycol. 2013;41:287–291.
- Bowman SM, Free SJ. The structure and synthesis of the fungal cell wall. Bioessays. 2006;28:799–808.
- Choi JW, Park HS. Improvement of Agrobacterium-mediated transformation of medicinal mushrooms. J Agric Life Sci. 2013;47:249–254.
- Muralidhar RV, Panda T. Fungal protoplast fusion - a revisit. Bioproc Eng. 2000;22:429–431.
- Chang ST, Li GSF, Peberdy JF. Isolation of protoplasts from edible fungi. World J Microbiol Biotechnol. 1985;1:185–193.
- Zhao J, Chang ST. Monokaryotization by protoplasting heterothallic species of edible mushrooms. World J Microbiol Biotechnol. 1993;9:538–543.
- Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes-applications to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2:113–118.
- Holland MJ, Holland JP. Isolation and identification of yeast messenger ribonucleic acids coding for enolase, glyceraldehyde-3-phosphate dehydrogenase, and phosphoglycerate kinase. Biochemistry. 1978;17:4900–4907.
- Hirano T, Sato T, Yaegashi K, et al. Efficient transformation of the edible basidiomycete Lentinus edodes with a vector using a glyceraldehyde-3-phosphate dehydrogenase promoter to hygromycin b resistance. Mol Gen Genet. 2000;263:1047–1052.
- Kuo CY, Chou SY, Huang CT. Cloning of glyceraldehyde-3-phosphate dehydrogenase gene and use of the gpd, promoter for transformation in Flammulina velutipes. Appl Microbiol Biotechnol. 2004;65:593–599.