Abstract
In the post-genomic era, gene function analysis has attracted much attention. Transformation is often needed to investigate gene function. In this study, an easy, rapid, reliable, and cost-effective colony polymerase chain reaction (PCR) method for screening mushroom transformants was developed: picking up a suitable amount of transformant’s tissue (1–10 μg) to 20 μl 0.25% Lywallzyme solution, and vortexing for 10 s followed by incubation at 34 °C for 15 min. Finally, 2 μl of the suspension was used as templates to perform PCR and single target bands were successfully amplified from respective transformants of Tremella fuciformis, Pleurotus ostreatus, and Pleurotus tuber-regium. This procedure could be widely employed for screening transformants in mushroom transformation experiments.
Keywords:
Mushrooms are filamentous macrofungi which have huge edible, medicinal, and economical value [Citation1]. Nowadays, more and more genome sequences of mushrooms have been completed [Citation2]. Gene functional analysis to facilitate mushroom biotechnology is an important task in the post-genomic era. Various reverse genetics approaches including gene silencing, gene targeting, and T-DNA insertional mutagenesis are available to investigate the gene function [Citation3–6]. Transformation is an important tool of reverse genetics research. However, screening many transformants by conventional PCR analysis is laborious and time-consuming because of the labor intensive process of DNA isolation [Citation7]. Because of this limitation, it is necessary to develop a rapid and effective colony PCR for screening transformants in mushroom transformation experiments.
Although direct colony PCR method was already established for fungi, several fungal species could not be successfully amplified [Citation8], pre-treatment of fungal colonies is still the most promising strategy [Citation9]. Novozym 234 was used for pre-treatment in lower fungi for colony PCR, but it needed additional incubation step for inactivation of enzyme [Citation10]. Commercial kits, like Ampdirect plus (Shimadzu, Kyoto, Japan) and Y-PER (Cat# 78990; Thermo Scientific, Waltham, MA) have been applied to colony PCR in bacteria, yeasts, and microalgae [Citation11,Citation12], but mushrooms were not tested extensively for successful colony PCR. With the assistance of microwave treatment, rapid and simple template preparation from colonies and fruit bodies in several mushrooms was successful for PCR amplification [Citation13]. However, thermal effects of microwave reduced the ability for amplification of large PCR products. Combining several kinds of pre-treatment together, a microwave, plMAN5C, proteinase K, and boiling (MMPB) method for colony PCR in red yeasts was developed [Citation14]. This method has several drawbacks including the time-consuming manipulation and the use of expensive reagents such as proteinase K.
In this study, a rapid and simple colony PCR was developed to screen transformants in several common mushrooms including Tremella fuciformis, Pleurotus ostreatus, and Pleurotus tuber-regium. These strains were preserved in the Laboratory of Food Microbiology, Huazhong Agricultural University. T. fuciformis and P. ostreatus were subcultured on potato dextrose agar (PDA) at 25 °C, P. tuber-regium at 32 °C. The plasmids carrying lac, hph, and egfp had been used and Agrobacterium tumefaciens-mediated transformation of these mushrooms was carried out as described before [Citation15–17].
For screening the transformants, a small amount of the cells (approximately, 1–10 μg, 0.5–1 mm3) or the actively growing edge of mycelia (1–10 μg and 1–2 mm in diameter) were picked up from the selective plates PDSA (50 μg/ml hygromycin B and 200 μg/ml cefotaxime) by using a sterilized toothpick and resuspended in 20 μl Lywallzyme solution (an effective cell wall lyticase, Guangdong Institute of Microbiology, Guangdong, China) in a 200 μl PCR tube. To facilitate the disruption of the tissue, the mixture was vortexed for 10 s and an additional incubation under 34 °C for 15 min. Vortexing can be omitted in some cases, especially for mycelia because of their characteristic of dispersion. A 2 μl suspension was used as template in PCR reactions. PCR reactions were performed in a 20 μl volume containing 2 μl 10 × EasyTaq buffer, 200 μM dNTPs, 0.2 μM of each primer, and 1 U EasyTaq DNA polymerase (TransGen Biotech, Beijing, China). The primers used in this study were listed in . The PCR procedures were: 95 °C for 3 min, followed by 34 cycles of denaturation at 95 °C for 30 s, annealing at 55–58 °C (according to different primers) for 30 s, extension at 72 °C and a final elongation step at 72 °C for 5 min. Ultrapure water was used instead of DNA templates in negative controls, while vectors were used as positive controls. The PCR products were examined by 1.0% (w/v) agarose gel electrophoresis.
Table 1. Primers used in this study.
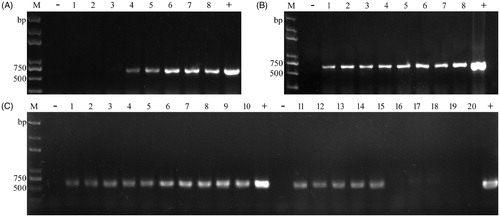
To make the procedure more cost-effective and convenient, we optimized it using transformants of T. fuciformis yeast-like cells (YLCs). Several important parameters such as concentration of enzyme, incubation time, quantity of template, and inactivation of enzyme were tested. The results showed lower concentration Lywallzyme (≤0.3125%) were capable of amplifying the desired fragment, higher concentration Lywallzyme (>0.3125%) failed to obtain the expected products (). A reasonable explanation is that PCR amplification was inhibited by higher concentration of Lywallzyme, while lower concentration of Lywallzyme has less inhibitory effect to PCR amplification and was enough to wound the cell wall of YLCs. We also explored whether the incubation time had influence on colony PCR in YLCs of T. fuciformis. The incubation time was varied from 15 to 120 min under the optimized enzyme concentration. The results revealed that it was not the most critical factor for PCR reactions, although longer time was beneficial for releasing DNA from cells (). In addition, the quantities of templates in supernatant (after centrifuging) was less than those in suspension, it had little influence in our protocols (). But inactivation of enzyme by heating treatment after incubation led to difficult PCR amplification. Distinctly weaker bands were obtained after 100 °C heating treatment compared to the untreated samples ().
Figure 1. Colony PCR amplification of hph from transformants of YLCs of T. fuciformis by changing parameters. (A) Under different concentrations of Lywallzyme treatment (2.5, 1.25, 0.625, 0.3125, 0.25, 0.125, 0.0625, and 0.03125%). 1–8: the same transformant; (B) Under the optimized enzyme concentration treatment for different times (15, 30, 45, 60, 75, 90, 105, and 120 min). 1–8: the same transformant; (C) Comparison between using the suspension and the supernatant both extracted under 0.25% Lywallzyme for 15 min; comparison between using the unheated suspension and the suspension followed by an additional boiling step at 100 °C for 5 min. The suspension was extracted under 0.25% Lywallzyme for 15 min. 1–5: using the supernatant as template. 6–10: using the suspension as template. 11–15: using the unheated suspension as template. 16–20: using the heated suspension as template. 1–10, 11–20: the same transformant. M: Trans2K plus DNA marker (Transgen, Beijing, China), −: negative control; +: positive control.
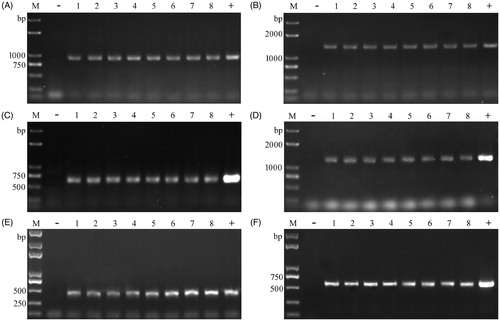
The optimal procedure was established: picking up a suitable amount of fungal tissue (approximately 1–10 μg) to 20 μl 0.25% Lywallzyme solution, vortexing for 10 s, and then incubation at 34 °C for 15 min, and 2 μl suspension was used as templates to perform PCR. It was used to screen transformants of T. fuciformis, P. ostreatus, and P. tuber-regium. Amplification of the lac3, hph, egfp, or egfp-hph was achieved successfully in transformants of these mushrooms after enzymolysis treatment ().
Figure 2. Colony PCR for screening transformants of T. fuciformis, L tuber-regium, and P. ostreatus. (A) for lac3-1 amplification in YLCs of T. fuciformis; (B) for lac3-2 amplification in YLCs of T. fuciformis; (C) for hph amplification in mycelia of T. fuciformis; (D) for egfp-hph cassette amplification in mycelia of T. fuciformis; (E) for egfp-1 amplification in L. tuber-regium; (F) for egfp-2 amplification in P. ostreatus. M: Trans2K plus DNA marker (Transgen, Beijing, China), −: negative control; +: positive control; 1–8: putative transformants.
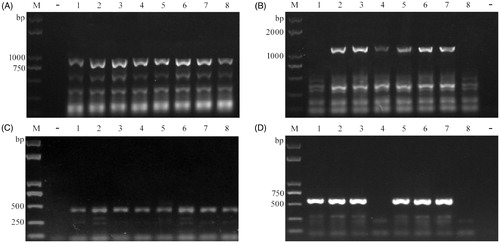
Compared with the conventional PCR analysis, the presented method requires neither the use of expensive and specialized equipment or toxic reagent like phenol used for DNA extraction. It significantly reduced the time and costs involved. In addition, in contrast to other published colony PCR protocols [Citation10,Citation11,Citation14], no additional boiling step was in general required for lysing the microorganisms or inactivating the enzyme. Its simplicity makes it convenient to prepare a large amount of samples at the same time. Compared to the current protocol [Citation13], our new method requires less harsh treatment of the sample and takes less time. A 5-minute incubation of cells and mycelia is sufficient to produce successful amplification using our colony PCR approach (data not shown). The operation steps of this method are simple, without the need for a cooling step after heat treatment. Using this procedure, more than 100 samples of mushroom DNA can be prepared in 30 min, whereas the microwave treatment method needs approximately 1 h. Noteworthy is that PCR products as large as ∼3 kb were obtained using this method (data not shown). Moreover, compared to a commercially available kit (2 × T5 Direct PCR kit) (Tsingke, Beijing, China), the electrophoresis results of this colony PCR was also satisfactory with specific amplification (). The addition of T5 DNA polymerase to the PCR system increased the rate of amplification compared to the EasyTaq DNA polymerase. However, potential primer bias existed in fungi and primer choice was critical for PCR success [Citation18,Citation19]. Multiple direct colony PCRs with different primer combinations or specific primers could be carried out to solve this problem [Citation20]. A suitable Lywallzyme concentration is also important for successful PCR amplification, in order to provide enough for cell breakage but not so much that it causes inhibition of the PCR reaction.
Figure 3. Colony PCR by using 2 × T5 direct PCR kit. (A) for lac3-1 amplification in YLCs of T. fuciformis; (B) for lac3-2 amplification in YLCs of T. fuciformis; (C) for egfp-1 amplification in L. tuber-regium; (D) for egfp-2 amplification in P. ostreatus. M: Trans2K plus DNA marker (Transgen, Beijing, China), −: negative control; +: positive control; 1–8: putative transformants.
In conclusion, a rapid and effective colony PCR procedure in several mushrooms was established. This method was really a useful tool to screen desired transformants from a large number of colonies at the same time.
Acknowledgments
The authors thank Daniel J. Ebbole, Professor in the Department of Plant Pathology and Microbiology at Texas A & M University for revising the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Mueller GM, Schmit JP, Leacock PR, et al. Global diversity and distribution of macrofungi. Biodivers Conserv. 2007;16:37–48.
- Li H, Wu S, Ma X, et al. The genome sequences of 90 mushrooms. Sci Rep. 2018;8:9982.
- Wälti MA, Villalba C, Buser RM, et al. Targeted gene silencing in the model mushroom Coprinopsis cinerea (Coprinus cinereus) by expression of homologous hairpin RNAs. Eukaryot Cell. 2006;5:732–744.
- Sato M, Kurahashi A, Nishibori K, et al. Development of a transformation system for the edible mushroom Grifola frondosa: demonstrating heterologous gene expression and RNAi-mediated gene silencing. Mycoscience. 2015;56:364–372.
- Hamer L, Adachi K, Montenegro-Chamorro MV, et al. Gene discovery and gene function assignment in filamentous fungi. Proc Natl Acad Sci USA. 2001;98:5110–5115.
- Salame TM, Knop D, Tal D, et al. Predominance of a versatile-peroxidase-encoding gene, mnp4, as demonstrated by gene replacement via a gene targeting system for Pleurotus ostreatus. Appl Environ Microbiol. 2012;78:5341–5352.
- van Burik JAH, Schreckhise RW, White TC, et al. Comparison of six extraction techniques for isolation of DNA from filamentous fungi. Med Mycol. 1998;36:299–303.
- Walch G, Knapp M, Rainer G, et al. Colony-PCR is a rapid method for DNA amplification of hyphomycetes. J Fungi. 2016;2:2–10.
- Pancher M, Ceol M, Corneo PE, et al. Fungal endophytic communities in Grapevines (Vitis vinifera L.) respond to crop management. Appl Environ Microbiol. 2012;78:4308–4317.
- Zeijl C, Kamp E, Punt PJ, et al. An improved colony-PCR method for filamentous fungi for amplification of PCR-fragments of several kilobases. J Biotechnol. 1997;59:221–224.
- Alshahni MM, Makimura K, Yamada T, et al. Direct colony PCR of several medically important fungi using Ampdirect® plus. Jpn J Infect Dis. 2009;62:164–167.
- Packeiser H, Lim C, Balagurunathan B, et al. An extremely simple and effective colony PCR procedure for bacteria, yeasts, and microalgae. Appl Biochem Biotechnol. 2013;169:695–700.
- Izumitsu K, Hatoh K, Sumita T, et al. Rapid and simple preparation of mushroom DNA directly from colonies and fruiting bodies for PCR. Mycoscience. 2012;53:396–401.
- Lin X, Yang F, Zhou Y, et al. Highly-efficient colony PCR method for red yeasts and its application to identify mutations within two leucine auxotroph mutants. Yeast. 2012;29:467–474.
- Ding Y, Liang S, Lei J, et al. Agrobacterium tumefaciens mediated fused egfp-hph gene expression under the control of gpd promoter in Pleurotus ostreatus. Microbiol Res. 2011;166:314–322.
- Zhu H, Liu D, Wang Y, et al. Use of the yeast-like cells of Tremella fuciformis as a cell factory to produce a Pleurotus ostreatus hydrophobin. Biotechnol Lett. 2017;39:1167–1173.
- Liu D, Zhu H, Chen Y, et al. Agrobacterium tumefaciens-mediated transformation of the king tuber medicinal mushroom Lentinus tuber-regium (Agaricomycetes). Int J Med Mushrooms. 2018;20:791–796.
- Bokulich NA, Mills DA. Improved selection of internal transcribed spacer-specific primers enables quantitative, ultra-high-throughput profiling of fungal communities. Appl Environ Microbiol. 2013;79:2519–2526.
- Bellemain E, Carlsen T, Brochmann C, et al. ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol. 2010;10:1–9.
- Wang YN, Liu XY, Zheng RY. Umbelopsis changbaiensis sp. nov. from China and the typification of Mortierella vinacea. Mycol Progress. 2014;13:657–669.
