Abstract
To explore species diversity of Hypocreaceae, collections from Guangdong, Hubei, and Tibet of China were examined and two new species and a new Chinese record were discovered. Morphological characteristics and DNA sequence analyses of the ITS, LSU, EF-1α, and RPB2 regions support their placements in Hypocreaceae and the establishments of the new species. Hypomyces hubeiensis sp. nov. is characterized by occurrence on fruitbody of Agaricus sp., concentric rings formed on MEA medium, verticillium-like conidiophores, subulate phialides, rod-shaped to narrowly ellipsoidal conidia, and absence of chlamydospores. Trichoderma subiculoides sp. nov. is distinguished by effuse to confluent rudimentary stromata lacking of a well-developed flank and not changing color in KOH, subcylindrical asci containing eight ascospores that disarticulate into 16 dimorphic part-ascospores, verticillium-like conidiophores, subcylindrical phialides, and subellipsoidal to rod-shaped conidia. Morphological distinctions between the new species and their close relatives are discussed. Hypomyces orthosporus is found for the first time from China.
1. Introduction
The family Hypocreaceae typified by Hypocrea Fr. was established by Saccardo [Citation1] and was redefined by Rossman et al. [Citation2] who treated it in a narrow sense and recognized 12 genera. Approximate 17 genera are currently accepted [Citation3–6], including Hypomyces (Fr.) Tul. & C. Tul and Trichoderma Pers., two major genera encompassing the majority of species of the family. The phylogenetic relationship among genera of the group was first revealed by Spatafora and Blackwell [Citation7].
This study is focused on two major genera of the family. For a long time, many fungicolous fungi with light- or bright-colored perithecia produced within subiculum were described as Hypomyces which is typified by H. lactifluorum (Schwein.) Tul. & C. Tul. Morphological characteristics and phylogenetic analysis based on sequence of nuclear ribosomal large subunit (LSU) rDNA in the previous work indicated that the genus is not a monophyletic group [Citation8–11]. After the comprehensive studies by Rogerson and Samuels [Citation12–15] and Põldmaa and collaborators [Citation11,Citation16–22], the generic concept of Hypomyces became clear. Among the 212 names listed in Index Fungorum database, about 77 species are commonly accepted [Citation11,Citation23–29]. Twenty-seven of them have been known from China [Citation27,Citation29–31]. Members of the genus are mainly distributed in temperate and tropical regions and economically important in biomedicine and agriculture [Citation32,Citation33].
Host specificity, color of subicula and perithecia, shape, size, septation, surface ornamentation, and apiculus of ascospores, and type of asexual states are main characters used for identifications of Hypomyces species. The genus grows on Agaricales, Boletales, Helotiales, and Pezizales are highly host-specific, while those occurring on Polyporales may have a slightly wider host range [Citation2,Citation12–16]. For example, H. lithuanicus Heinr.-Norm. lives only on Lactariustor minosus (Schaeff.) Gray, H. hyalinus (Schwein.) Tul. & C. Tul. is restricted to Amanita Adans, and H. melanocarpus Rogerson & Mazzeris is on Tylopilus P. Karst. However, H. australis (Mont.) Höhn., H. rosellus (Alb. & Schwein.) Tul. & C. Tul. and H. tegillum Berk. & M.A. Curtis show the least specialized parasites and even being found on non-fungus substrates, like rotten bark and wood [Citation14].
Trichoderma, the largest genus in the family Hypocreaceae, was originally established by Persoon [Citation34] and typified with T. viride Pers. Since then, number of Trichoderma species increased dramatically. Bissett et al. [Citation35] provided a list of 254 Trichoderma species with DNA sequences or living cultures available. There are more than 340 species currently recognized in the genus. They occur on rotten wood, bark and leaves, fruitbodies of other fungi, in soil, or within healthy plant tissues as endophytes [Citation36–39]. They are renewable natural resources and play important roles in production of industrial enzymes and antibiotics [Citation40], biological control of soil-borne plant pathogens [Citation41,Citation42], plant growth promotion [Citation43], induction of plant resistance [Citation44], production of bioactive secondary metabolites [Citation36] and remediation of soil contaminated by heavy metals [Citation45]. Taxonomy of Trichoderma species is mainly based on anatomy of stromata and perithecia, color, shape, size of ascospores, conidia and chlamydospores, type of conidiophores, colony morphology and growth rate, and DNA sequence data [Citation46,Citation47]. The phylogenetic analyses based on translation elongation factor 1-α encoding (EF-1α) and RNA polymerase II subunit 2 (RPB2) regions indicated that the hyaline ascospored species are divided into 11 clades and those of green ones are separated into 7 clades [Citation48–50].
During our survey of hypocrealean species on fungi and plant debris in China, two undescribed taxa are found based on morphological characteristics and DNA sequence analyses of the internal transcribed spacer (ITS), LSU, EF-1α, and RPB2. Differences between the new species and their close relatives are discussed. Hypomyces orthosporus is reported for the first time from China.
2. Materials and methods
2.1. Collections and morphological study
Specimens were collected from Shennongjia Forestry District of Hubei and Chebaling National Nature Reserve of Guangdong and Mainling of Tibet, and deposited in the Herbarium Mycologicum Academiae Sinicae (HMAS). Cultures are kept in the State Key Laboratory of Mycology, Institute of Microbiology, Chinese Academy of Sciences. Methods used by Jaklitsch [Citation46] and Põldmaa [Citation19] were followed. Test for color change of perithecial wall was made with 3% potassium hydroxide (KOH). To observe anatomic structures of perithecia, longitudinal sections of ascomata were made with a freezing microtome (YD-1508-III; Jinhua Yidi Medical Appliance Co., Jinhua, China) at a thickness of 6–8 µm. Microscopic examinations and measurements were taken from the sections and squash mounts in lactophenol cotton blue solution using an Olympus BH-2 microscope (Tokyo, Japan). Photographs were taken with a Leica DFC450 digital camera (Leica Camera, Wetzlar, Germany) attached to a Leica M125 stereomicroscope (Leica) for gross morphology and a Zeiss AxioCamMRc 5 digital camera (Carl Zeiss, Jena, Germany) attached to a Zeiss Axio Imager A2 microscope (Carl Zeiss) for anatomic structures. Measurements of individual structures were based on 30 units, except when otherwise noted. Cultures were obtained from conidia on subiculum or from fresh ascomata using single ascospore isolation. To determine colony features and growth rates, strains were grown on cornmeal dextrose agar (CMD; Yuanye Bio-Technology Co. Ltd., Shanghai, China), malt extract agar (MEA; Oxoid Ltd., Basingstoke, UK), potato dextrose agar (PDA; HuiXing Biochemistry Reagent Ltd Co., Shanghai, China) and synthetic nutrient-poor agar (SNA) [Citation51] in 90 mm plastic Petri dishes at 25 °C for 7 or 14 d. For observation of conidiophores and microconidia, cultures were grown on SNA at 25 °C with alternating periods of light and darkness (12 h/12 h).
2.2. DNA extraction, PCR amplification, and sequencing
The genomic DNA was extracted from fresh mycelium following the methods of Wang and Zhuang [Citation52]. Primer pairs, ITS5/ITS4 [Citation53], LR0R/LR5 [Citation9, Citation54], and EF1-728F/EF1567R [Citation55,Citation56] were used to amplify the sequences of ITS, LSU, and EF-1α regions for Hypomyces species, while EF1-728F/TEF1LLErev [Citation55,Citation57] and fRPB2-5F/fRPB2-7cR [Citation58] were applied to amplify the sequences of EF-1α and RPB2 regions for Trichoderma species. PCR reactions were performed on an ABI 2720 Thermal Cycler (Applied Biosciences, Foster City, CA) with a 25 µl reaction system consisting of 12.5 µl Taq MasterMix, 1 µl each primer (10 µM), 1 µl template DNA and 9.5 µl ddH2O, based on the procedures detailed in White et al. [Citation53], Chaverri and Samuels [Citation59], Rehner and Buckley [Citation56], and Liu et al. [Citation58]. DNA sequencing was carried out in both directions on an ABI 3730XL DNA Sequencer (Applied Biosciences).
2.3. Sequence alignment and phylogenetic analyses
Newly generated sequences and those retrieved from GenBank are listed in (Hypomyces) and (Trichoderma). Nectria eustromatica Jaklitsch & Voglmayr and Thyronectria berolinensis (Sacc.) Seaver were used as outgroup taxa. Sequences were assembled, aligned, and the primer sequences were trimmed with BioEdit version 7.0.5 (Ibis Biosciences, Carlsbad, CA) [Citation60] and converted to NEXUS files by ClustalX version 1.83 (EMBL, Heidelberg, Germany) [Citation61]. Sequences were first subjected to the BLAST searches to determine preliminarily their taxonomic positions. TrichOKEY [Citation62] was also applied to preliminary identification of Trichoderma. Due to a low number of variable sites and long insertions in certain species of Trichoderma [Citation63], ITS sequences were not incorporated into phylogenetic analyses. The partition homogeneity test of ITS, LSU, and EF-1α sequences of Hypomyces, EF-1α and RPB2 sequences of Trichoderma were performed with PAUP version 4.0b10 (Sinauer Associates, Sunderland, MA) [Citation64]. To confirm the phylogenetic positions of the new species, sequences of these regions were combined and analyzed with maximum parsimony (MP) and maximum likelihood (ML) methods. The MP analysis was performed with PAUP version 4.0b10 [Citation64] using 1000 replicates of heuristic search with random addition of sequences and subsequent TBR (tree bisection and reconnection) branch swapping. Topological confidence of resulted trees was tested by maximum parsimony bootstrap proportion (MPBP) with 1000 replications, each with 10 replicates of random addition of taxa. Four Markov chains were run simultaneously for 1,000,000 generations with the trees sampled every 100 generations. A 50% majority rule consensus tree was computed after excluding the first 2500 trees as “burn-in.” The ML analysis was conducted with IQ-Tree version 1.6.10 (University of Vienna, Vienna, Austria) [Citation65] using the best model for each locus chose by ModelFinder [Citation66]. Branch support measures were calculated with 1000 bootstrap replicates. Trees were examined via TreeView version 1.6.6 (University of Glasgow, Glasgow, UK) [Citation67]. Maximum likelihood bootstrap (MLBP) and MPBP greater than 50% are shown at the nodes.
Table 1. List of Hypomyces species, herbarium/strain numbers and GenBank accession numbers of materials used in this study.
Table 2. List of Hypocreaceae species, herbarium/strain numbers and GenBank accession numbers of materials used in this study.
3. Results
3.1. Phylogenetic analyses
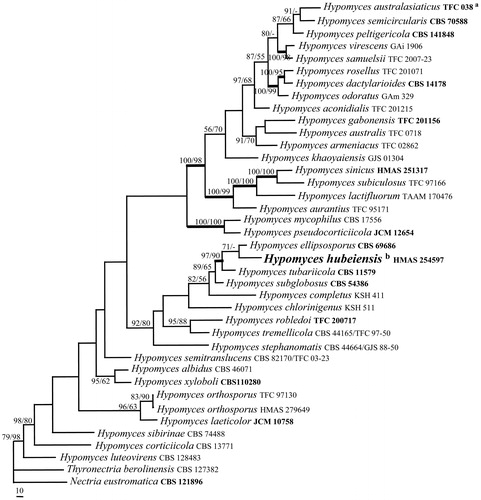
To determine the positions of the Hypomyces collections, the sequences of ITS, LSU, and EF-1α regions of 36 Hypomyces species were analyzed. The PHT (p=.01) indicated that the individual partitions were not highly incongruent [Citation68], thus the three loci were thus combined for phylogenetic analyses. The combined datasets include 2440 characters, of which 1704 were constant, 233 variable and parsimony-uninformative and 503 parsimony-informative. The MP analysis resulted in a single most parsimonious tree (tree length = 2459, CI = 0.4429, HI = 0.5571, RI = 0.5287, RCI = 0.2342). The final matrix was deposited in TreeBASE with accession No. S23771. The MP tree generated is shown in . The topology of the ML tree is similar to that of the MP tree. HMAS 254597 clustered with representative species of Hypomyces (MLBP/MPBP = 79%/98%), which confirmed its taxonomic position in the genus.
Figure 1. A MP tree generated based on the combined datasets of ITS, LSU and EF-1α sequences of Hypomyces species. Supporting values showing at branches: MLBP (left) and MPBP (right). MLBP and MPBP greater than 50% are shown at the nodes. The branch support values ≥90 are indicated by thicker lines. Genbank accession numbers in bold indicate the sequences from ex-type strains. The scale bars indicate number of nucleotide substitutions per site.
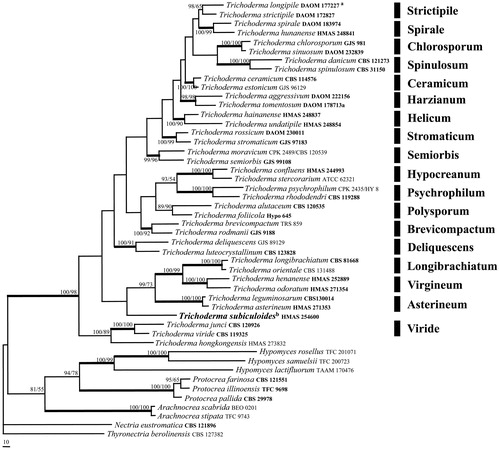
To place the Trichoderma collection, the sequences of EF-1α and RPB2 regions from 38 species representing 18 clades of Trichoderma, three species of Protocrea, three of Hypomyces and two of Arachnocrea were analyzed by the methods of ML and MP. The PHT (p=.01) indicated that the individual partitions were not highly incongruent [Citation68], the two loci were thus combined for phylogenetic analyses. The combined datasets include 1698 characters, of which 1027 were constant, 118 variable and parsimony-uninformative and 553 parsimony-informative. The MP analysis resulted in three most parsimonious trees (tree length = 3602, CI = 0.2984, HI = 0.7016, RI = 0.5171, RCI = 0.1543). The final matrix was deposited in TreeBASE with accession No. S23772. One of the three MP trees generated is shown in . The ML tree is of a similar tree topology. HMAS 254600 was shown as a separate lineage associated with Asterineum, Longibrachiatum, and Virgineum clades of Trichoderma, and further clustered with other species of the genus forming a highly supported monophyletic group (MLBP/MPBP = 100%/98%), which confirmed its taxonomic position in the genus.
Figure 2. A MP tree generated based on the combined datasets of EF-1α and RPB2 sequences of Trichoderma species and relatives. Supporting values showing at branches: MLBP (left) and MPBP (right). MLBP and MPBP greater than 50% are shown at the nodes. The branch support values ≥90 are indicated by thicker lines. Genbank accession numbers in bold indicate the sequences from ex-type strains. The scale bars indicate number of nucleotide substitutions per site.
3.2. Taxonomy
3.2.1. Hypomyces hubeiensis Z.Q. Zeng & W.Y. Zhuang, sp. nov.
Fungal Names: FN570597.
Description: On CMD, colony radius 14 mm after 7 d at 25 °C, velvet, producing yellowish green pigment in medium, reverse yellowish green; aerial hyphae white, scarce. On MEA, colony radius 13 mm after 7 d at 25 °C, velvet, surface white, reverse white; aerial hyphae white, scarce, forming concentric rings. On PDA, colony radius 13 mm after 7 d at 25 °C, floccose, surface grey white, reverse light sienna; aerial hyphae white, dense, floccose. Conidiophores arising from aerial hyphae, branched, septate, 1–2-verticillate, with terminal whorl of 2–6 phialides. Phialides subulate, tapering toward apex, smooth, 8–20 × 2–3 µm. Conidia rod-shaped to narrowly ellipsoidal, aseptate, hyaline, smooth, 3–6 × 1–2.3 µm.
Etymology: The specific epithet refers to the type locality.
Holotype: China, Hubei Province, Shennongjia Forestry District, Dajiuhu, on Agaricus sp., September 17 2014, Z.Q. Zeng, W.T. Qin, K. Chen & H.D. Zheng 9791 (HMAS 254597) ().
Figure 3. Hypomyces hubeiensis (HMAS 254597). (A−C) Cultures after 14 d at 25 °C (A: on CMD, B: on MEA, C: on PDA); (D–I) Conidiophores, phialides, and conidia; (J) Phialides and conidia; (K–M) onidia. Scale bar = 10 μm.
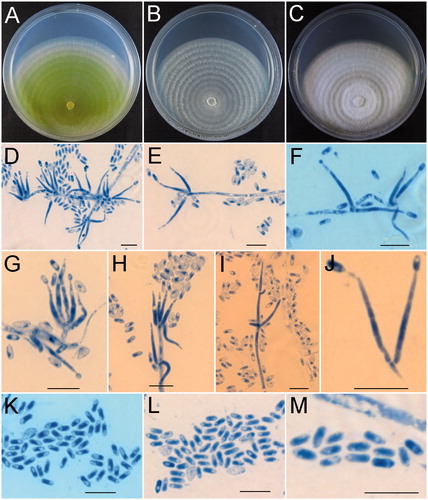
Notes: The new species grows on fruitbodies of Agaricus sp. containing only the asexual state. Among the known agaricicolous species of Hypomyces, H. hubeiensis is morphologically similar to H. succineus Rogerson & Samuels and H. tremellicola (Ellis & Everh.) Rogerson in forming verticillium-like conidiophores. But H. succineus differs in occurring on Pholiota sp. rather than Agaricus sp. and having much larger conidia [(7–)8.8–13.3(–16)×(2.4–)3.3–4.2(–5) µm vs. 3–6 × 1–2.3 µm] [Citation15]. H. tremellicola grows on Crepidotus spp. and has larger conidia [5–9 × 3–4(–5) µm vs. 3–6 × 1–2.3 µm] [Citation15].
3.2.2. Hypomyces orthosporus K. Põldmaa
Mycotaxon 59: 390 (1996)
= Cladobotryum orthosporum (W. Gams) K. Põldmaa, Mycotaxon 59: 390 (1996)
≡ Sibirina orthospora W. Gams, Persoonia 7: 163 (1973)
On CMD, colony radius 46 mm after 7 d at 25 °C, floccose, producing light yellowish brown pigment, reverse brown; aerial hyphae white, scarce. On MEA, colony radius 40 mm after 7 d at 25 °C, floccose, producing light yellowish brown pigment, reverse brown; aerial hyphae white, scarce. On PDA, colony radius 42 mm after 7 d at 25 °C, floccose, producing light yellowish brown pigment; aerial hyphae white, scarce. Conidiophores arising from aerial mycelium, indefinite in length, 1–2-verticillate, with terminal whorl of 3–10 phialides. Phialides subulate, tapering toward apex, hyaline, smooth, 10–35 µm long, 1–1.8 µm at the base. Conidia subcylindrical, sometimes subfusiod, rarely narrowly ellipsoidal (0–)1(–2)-septate, hyaline, smooth, with a rounded tip and a basal hilum, 10–18 × 2.5–5 µm.
Specimen examined: China, Tibet, Nyingchi, Mainling, alt. 2800 m, on fruiting body of a polypore, September 12 2016, H.D. Zheng, Z.Q. Zeng, X.C. Wang, K. Chen & Y.B. Zhang 10736 (HMAS 279649) ().
Figure 4. Hypomyces orthosporus (HMAS 279649). (A−C) Cultures after 7 d at 25 °C (A: on CMD, B: on MEA, C: on PDA); (D) Conidiophores and phialides; (E−G) Phialides and conidia; (H−L) onidia. Scale bar: D−G = 10 μm; H−L = 5 μm.
Known distribution: China, Estonia, Finland, and The Netherlands.
Notes: Sibirina orthospora was described by Gams [Citation69] based on the specimen on decaying wood from The Netherlands with only asexual state described. The sexual and asexual stage connection of the fungus was established by Poldmaa [Citation16] based on the materials collected from Estonia. The phylogenetic tree based on LSU sequences showed that the Chinese collection (HMAS 279649) associated with that from Estonia (TFC 97-130) receiving high support values (MLBP/MPBP = 83%/90%).
3.2.3. Trichoderma subiculoides Z.Q. Zeng & W.Y. Zhuang, sp. nov
Fungal Names: FN570596.
Description: Stromata broadly attached on natural substratum, widely effuse to confluent, rudimentary and somewhat subiculum-like, lacking of a defined margin or flank, whitish to beige when dry, cinnamon brown after rehydration, not changing color in 3% KOH, 3–7 × 2–5 mm, 0.4 mm thick. Ostiolar dots distinct, dirty brownish to light brown when dry, brown to dark brown when rehydrated. In section, cortical tissue of textura globulosa, 5–25 µm thick, cells light yellow, 1.5–5 × 1.5–4.5 µm; subcortical tissue of textura intricata, hyphae hyaline to pale brown, 2–3.5 µm thick; subperithecial tissue of textura epidermoidea, cells hyaline, thin-walled, 5–10 × 3–5 µm. Perithecia globose, subglobose to pyriform, 138–193 × 105–150 µm; peridium 6–12 µm thick at flanks, 15–30 µm thick at the base. Papilla prominent, blunt or truncate, brown, 18–63 µm high, 35–58 µm wide at the base. Asci subcylindrical, 78–115 × 2.8–5 µm. Part-ascospores hyaline, smooth, dimorphic, distal cells broadly ellipsoidal to globose, 3.5–5 × 2–4 µm, l/w 1–2; proximal cells ellipsoidal, 4–5 × 2–4 µm, l/w 1.3–2.
On CMD, colony radius 10 mm after 7 d at 20 °C, 41 mm at 25 °C, no growth at 30 and 35 °C, white, velvet; aerial hyphae scarce, hyaline. On PDA, colony radius 10 mm after 7 d at 20 °C, 26 mm at 25 °C, 8 mm at 30 °C, no growth at 35 °C, white, velvet; aerial hyphae dense, hyaline. On SNA, colony radius 9 mm after 7 d at 20 °C, 5 mm at 25 °C, no growth at 30 and 35 °C, producing cream to pale yellow pigment; aerial hyphae hyaline, scarce. Conidiophores arising from aerial mycelium, branched, branches septate, 1–2-verticillate, with the terminal whorl of 2–4 phialides, 15–55 × 2–3.5 µm. Phialides subcylindrical, tapering toward apex, smooth, 5–25 × 1.5–3 µm. Conidia subellipsoidal to rod-shaped, hyaline, smooth, 3–9 × 1.5–3 µm. No distinct odor detected.
Etymology: The specific epithet refers to the subiculum-like and rudimentary stromata.
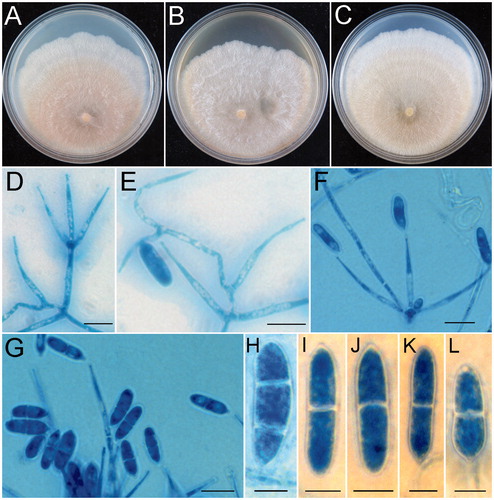
Holotype: China, Guangdong Province, Shixing County, Chebaling National Nature Reserve, on rotten branch, 2 November 2015, Z.Q. Zeng, X.C. Wang, K. Chen & Y.B. Zhang 10623 (HMAS 254600) ().
Figure 5. Trichoderma subiculoides (HMAS 254600). (A,B) Stroma on nature substrate; (C) Color of stroma after rehydration; (D) Color of rehydrated stroma in 3% KOH; (E−G) Cultures after 14 d at 25 °C (E: on CMD, F: on SNA, G: on PDA); (H) Perithecium in section; (I) Structure of perithecial at upper portion; (J) Ascus with ascospores; (K−O) Part-ascospores; (P) Phialides and conidia; (Q) Cortical and subcortical tissues in section; (R) Subperithecial tissue in section; (S−W) Conidia. Scale bars: A = 1 cm; B−D = 1 mm; H, I = 50 μm; J−P = 10 μm; Q, R = 20 μm; S−W = 10 μm.
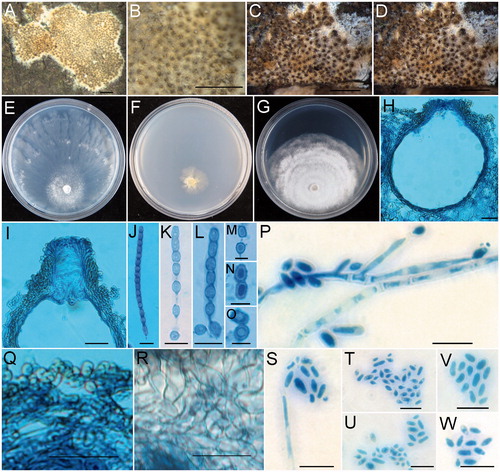
Notes: Among the known species of Trichoderma, T. subiculoides is morphologically similar to T. confluens W.T. Qin & W.Y. Zhuang and T. pseudolacteum C.S. Kim & N. Maek. in having effuse to confluent stromata which are broadly attached to substrates [Citation70,Citation71]. However, T. subiculoides differs from T. confluens in stromatal gross morphology and perithecia not changing color in 3% KOH, smaller perithecia (138–193 × 105–150 µm vs. 180–268 × 123–185 µm), ellipsoidal instead of globose, subglobose to nearly wedge-shaped proximal part-ascospores, and the absence of chlamydospores [Citation70]. The RPB2 sequence of T. subiculoides differs from that of T. confluens by 64 bp divergences in a total length of 751 bp. Trichoderma subiculoides can be easily distinguished from T. pseudolacteum by narrower asci (2.8–5 µm vs. 5.9–7.1 µm wide), smaller part-ascospores (distal 3.5–5 × 2–4 µm vs. 5.4–6.5 × 5.0–5.9 µm, proximal 4–5 × 2–4 µm vs. 5.3–6.9 × 4.3–5.2 µm), ellipsoidal to rod-shaped rather than globose to subglobose conidia [Citation71]. Sequence comparisons revealed that there are 62 bp unmatched loci among 452 bp for partial RPB2 region between the type strains (HMAS 254600 and TUFC 61490).
4. Discussion
Hypomyces is connected with diverse asexual states, such as mycogone-like, stephanoma-like, papulaspora-like, sepedonium-like, verticillium-like, acremonium-like, and cladobotryum-like [Citation2]. Host fungi in combination with types of asexual states are regarded as important taxonomic criteria for species identifications [Citation17]. Asexual states are sometimes even critical to distinguish genera in Hypocreaceae. Due to that H. berkeleyanus Plowr. & Cooke and H. broomeanus Tul. & C. Tul. are of gliocladium-like asexual states, Rehner and Samuels [Citation9] and Põldmaa et al. [Citation11] excluded them from Hypomyces and transferred these two species to another genus Sphaerostilbella (Henn.) Sacc. & D. Sacc. The taxonomic position of H. hubeiensis is revealed based on the sequence analyses of ITS, LSU, and EF-1α regions () as well as the morphological characters such as the substrate and verticillate conidiophores. However, its complete life cycle and infra-specific variation await future investigation. Some Hypomyces species with asexual states unknown due to their ascospores not germinating in laboratory condition [Citation27]. Establishment of asexual and sexual state connections for these fungi will provide essential information about life cycle of the whole fungus.
Stroma is a vegetative tissue that subtends or surrounds the ascomata [Citation2]. Tissues of the stromata of Trichoderma are composed of textura angularis, textura globulosa, textura intricata, textura prismatica, and textura epidermoidea depending on locations of the tissues. Stromatal anatomy is considered as one of the important morphological characters at generic and species levels for taxonomy of Hypocreaceae [Citation2]. Most species of Trichoderma have pulvinate, disciform, flat, peltate, turbinate, hemisphericalor or clavate stromata which are usually of a well-defined margin and flanks [Citation2,Citation59,Citation72], while very few species possess rudimentary or subiculum-like stromata, such as T. alcalifuscescens (Overton) Jaklitsch & Voglmayr, T. delicatulum Jaklitsch and T. parmastoi (Overton) Jaklitsch & Voglmayr [Citation47]. In Hypocreaceae the genera Arachnocrea Z. Moravec and Protocrea Petch produce subiculum surrounding their perithecia. However, the anatomic structure of T. subiculoides reveals that the fungus has a subiculum-like stroma instead of true subiculum that a cortical layer is present on the stromatal upper surface and the individual stromata lack of well-developed margin and flanks. Sequence analyses indicated that T. subiculoides clustered with other Trichoderma species receiving high bootstrap supports (MLBP/MPBP = 100%/98%) as a separate lineage and does not belong to any existing clades.
Acknowledgments
The authors are very grateful to Drs. H.D. Zheng, X.C. Wang, W.T. Qin, K. Chen, and Y.B. Zhang for collecting specimens jointly for this study, also to Dr. J.X. Deng for useful suggestion and to Mr. J. Fan for technical help.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Saccardo PA. Sylloge Fungorum II. Padova, Italy: Saccardo; 1883.
- Rossman AY, Samuels GJ, Rogerson CT, et al. Genera of Bionectriaceae, Hypocreaceae and Nectriaceae (Hypocreales, Ascomycetes). Stud Mycol. 1999;42:1–260.
- Maharachchikumbura SSN, Hyde KD, Jones EBG, et al. Towards a natural classification and backbone tree for Sordariomycetes. Fungal Divers. 2015;72:199–301.
- Maharachchikumbura SSN, Hyde KD, Jones EBG, et al. Families of Sordariomycetes. Fungal Divers. 2016;79:1–317.
- Zeng ZQ, Zhuang WY. Phylogenetic position of Pseudohypocrea (Hypocreales). Mycoscience. 2017;58:274–281.
- Wijayawardene NN, Hyde KD, Lumbsch HT, et al. Outline of Ascomycota: 2017. Fungal Divers. 2018;88:167–263.
- Spatafora JW, Blackwell M. Molecular systematic of unitunicate perithecial ascomycetes: the Clavicipitales-Hypocreales connection. Mycologia. 1993;85:912–922.
- Seifert KA. A monograph of Stilbella and some allied hyphomycetes. Stud Mycol. 1985;27:1–235.
- Rehner SA, Samuels GJ. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol Res. 1994;98:625–634.
- Candoussau F, Magni JF. New French records. Mycologist. 1995;9:12–14.
- Põldmaa K, Larsson E, Kõjalg U. Phylogenetic relationships in Hypomyces and allied genera, with emphasis on species growing on wood-decaying homobasidiomycetes. Can J Bot. 1999;88:1756–1768.
- Rogerson CT, Samuels GJ. Species of Hypomyces and Nectria occurring on Discomycetes. Mycologia. 1985;77:763–783.
- Rogerson CT, Samuels GJ. Boleticolous species of Hypomyces. Mycologia. 1989;81:413–432.
- Rogerson CT, Samuels GJ. Polyporicolous species of Hypomyces. Mycologia. 1993;85:231–272.
- Rogerson CT, Samuels GJ. Agaricolous species of Hypomyces. Mycologia. 1994;86: 839–866.
- Põldmaa K. A new species of Hypomyces and three of Cladobotryum from Estonia. Mycotaxon. 1996;59:389–405.
- Põldmaa K. Generic delimitation of the fungicolous Hypocreaceae. Stud Mycol. 2000;45:83–94.
- Põldmaa K. Three species of Hypomyces growing on basidiomata of Stereaceae. Mycologia. 2003;95:921–933.
- Põldmaa K. Tropical species of Cladobotryum and Hypomyces producing red pigments. Stud Mycol. 2011;68:1–34.
- Põldmaa K, Samuels GJ. Aphyllophoricolous species of Hypomyces with KOH negative perithecia. Mycologia. 1999;91:177–199.
- Põldmaa K, Samuels GJ. Fungicolous hypocreaceae (Ascomycota: Hypocreales) from Khao Yai National Park, Thailand. Sydowia. 2004;56:79–130.
- Põldmaa K, Lodge DJ, Samuels GJ. Three new polyporicolous species of Hypomyces and their Cladobotryum anamorphs. Sydowia. 1997;49:80–93.
- Kirk PM, Cannon PF, Minter DW, et al. Dictionary of the fungi. 10th ed. Wallingford, England: CABI; 2008.
- Zhuang WY, Chen SL, Zeng ZQ, et al. A new species of Hypomyces (Hypocreales) on Schizophyllum sp. from China. Mycosystema. 2012;31:821–826.
- Rossman AY, Seifert KA, Samuel GJ, et al. Genera in Bionectriaceae, Hypocreaceae, and Nectriaceae (Hypocreales) proposed for acceptance or rejection. IMA Fungus. 2013;4:41–51.
- Zare R, Gams W. More white verticillium-like anamorphs with erect conidiophores. Mycol Progress. 2016;15:993–1030.
- Zeng ZQ, Zhuang WY. Three new species and two new Chinese records of Hypomyces (Hypocreales). Mycosystema. 2016;35:1048–1055.
- Lechat C, Gardiennet A, Fournier J. First report of a lichenicolous species of Hypomyces (Hypocreaceae), H. peltigericola sp. nov. Ascomycete.org. 2017;9:23–26.
- Wei IC, Kirschner R. Two fungicolous anamorphic species of Hypomyces s. lat. from Taiwan. Fung Sci. 2017;32:15–25.
- Teng SC. Fungi of China. Beijing, China: Science Press; 1963.
- Wang YZ, Wu SH, Chou WN, et al. List of the Fungi in Taiwan. Taipei, Taiwan: Committee for Agriculture; 1999.
- Tamm H, Põldmaa K. Diversity, host associations, and phylogeography of temperate aurofusarin-producing Hypomyces/Cladobotryum including causal agents of cobweb disease of cultivated mushrooms. Fungal Biol. 2013;117:348–367.
- Liu L, Liu RX, Basnet BB, et al. A new seco-pimarane diterpene and four new β-resorcylic acid lactones from a fungicolous Hypomyces subiculosus. RSC Adv. 2017;7:51986–51992.
- Persoon CH. Dispositio methodica fungorum. Romer’s Neues Magazin Botanische. 1794;1:81–128.
- Bissett J, Gams W, Jaklitsch W, et al. Accepted Trichoderma names in the year 2015. IMA Fungus. 2015;6:263–295.
- Mukherjee PK, Horwitz BA, Singh US, et al. Trichoderma: biology and applications. Wallingford, England: CABI; 2013.
- Chen K, Zhuang WY. Seven soil-inhabiting new species of the genus Trichoderma in the Viride clade. Phytotaxa. 2017;312:28–46.
- Zhang YB, Zhuang WY. Two new species of Trichoderma with green ascospores on woody substrates. Mycosystema. 2019;38:11–22.
- Zhang YB, Zhuang WY. Epitypifications of three Trichoderma species and new Chinese records of the genus. Mycosystema. 2019;38:23–38.
- Jangir M, Pathak R, Sharma S. Trichoderma and its potential applications. In: Singh D, Singh H, Prabha R, editors. Plant-microbe interactions in agro-ecological perspectives. Singapore: Springer; 2017.
- Hanada RE, Jorge Souza TD, Pomella AW, et al. Trichoderma martiale sp. nov., a new endophyte from sapwood of Theobroma cacao with a potential for biological control. Mycol Res. 2008;112:1335–1343.
- Cheng CH, Yang CA, Peng KC. Antagonism of Trichoderma harzianum ETS 323 on Botrytis cinerea mycelium in culture conditions. Phytopathology. 2012;102:1054–1063.
- Stewart AHR. Applications of Trichoderma in plant growth promotion. In: Gupta VK, Schmoll M, Herrera-Estrella A, Upadhyay RS, Druzhinina I, Tuohy M, editors. Biotechology and biology of Trichoderma. Netherlands: Elsevier; 2014. p. 415–428.
- Harman GE. Myths and dogmas of biocontrol. Changes in perceptions derived from research on Trichoderma harzianum T22. Plant Dis. 2000;84:377–393.
- Joshi PK, Swarup A, Maheshwari S, et al. Bioremediation of heavy metals in liquid media through fungi isolated from contaminated sources. Indian J Microbiol. 2011;51:482–487.
- Jaklitsch WM. European species of Hypocrea. Part I. The green-spored species. Stud Mycol. 2009;63:1–91.
- Jaklitsch WM. European species of Hypocrea Part II: species with hyaline ascospores. Fungal Divers. 2011;48:1–250.
- Jaklitsch WM, Voglmayr H. Biodiversity of Trichoderma (Hypocreaceae) in Southern Europe and Macaronesia. Stud Mycol. 2015;80:1–87.
- Qin WT, Zhuang WY. Four new species of Trichoderma with hyaline ascospores from central China. Mycol Prog. 2016;15:811–825.
- Zhang YB, Zhuang WY. Four new species of Trichoderma with hyaline ascospores from southwest China. Mycosphere. 2017;8:1914–1929.
- Nirenberg HI. Studies on the morphologic and biologic differentiation in Fusarium section Liseola. Mitt Biol Bundesanst Land Forstwirtsch. 1976;169:1–117.
- Wang L, Zhuang WY. Designing primer sets for amplification of partial calmodulin genes from penicillia. Mycosystema. 2004;23:466–473.
- White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York (NY): Academic Press; 1990. p. 315–322.
- Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172:4238–4246.
- Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–556.
- Rehner SA, Buckley E. A Beauveria phylogeny inferred from nuclear ITS and EF1-alpha sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 2005;97:84–98.
- Jaklitsch WM, Komon M, Kubicek CP, et al. Hypocrea voglmayrii sp. nov. from the Austrian Alps represents a new phylogenetic clade in Hypocrea/Trichoderma. Mycologia. 2005;97:1365–1378.
- Liu YJ, Whelen S, Hall BD. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol Biol Evol. 1999;16:1799–1808.
- Chaverri P, Samuels GJ. Hypocrea/Trichoderma (Ascomycota, Hypocreales, Hypocreaceae): species with green ascospores. Stud Mycol. 2003;48:1–116.
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98.
- Thompson JD, Gibson TJ, Plewniak F, et al. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882.
- Druzhinina IS, Kopchinskiy AG, Komoń M, et al. An oligonucleotide barcode for species identification in Trichoderma and Hypocrea. Fungal Genet Biol. 2005;42:813–828.
- Samuels GJ, Dodd S, Lu BS, et al. The Trichoderma koningii aggregate species. Stud Mycol. 2006;56:67–133.
- Swofford DL. PAUP 4.0b10: phylogenetic analysis using parsimony. Sunderland (MA): Sinauer Associates; 2002.
- Nguyen LT, Schmidt HA, von Haeseler A, et al. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol. 2015;32:268–274.
- Chernomor O, von Haeseler A, Minh BQ. Terrace aware data structure for phylogenomic inference from supermatrices. Syst Biol. 2016;65:997–1008.
- Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358.
- Cunningham CW. Can three incongruence tests predict when data should be combined? Mol Biol Evol. 1997;14:733–740.
- Gams W. Phialides with solitary conidia? Persoonia. 1973;7:161–169.
- Qin WT, Zhuang WY. Two new hyaline-ascospored species of Trichoderma and their phylogenetic positions. Mycologia. 2016;108:205–214.
- Kim CS, Shirouzu T, Nakagiri A, et al. Trichoderma eijii and T. pseudolacteum, two new species from Japan. Mycol Prog. 2013;12:739–753.
- Zhu ZX, Zhuang WY. Three new species of Trichoderma with hyaline ascospores from China. Mycologia. 2015;107:328–345.
