Abstract
Invasive fungal infections caused by Cyberlindnera fabianii have recently increased. However, biochemical kits such as API 20 C AUX and Vitek-2C have misidentified this species as other Candida spp. such as C. pelliculosa or C. utilis due to no information of Cy. fabianii in yeast database. During our 2016–2017 surveys, eleven isolates of Cy. fabianii were obtained in International St. Mary’s Hospital in Korea. Here, we describe its morphological and molecular characteristics and tested its antifungal susceptibility against nine antifungal agents. The sequences of the ITS region and the D1/D2 region of LSU revealed 100% identity with the sequences of Cy. fabianii. In comparison with the results from MALDI-TOF mass spectrometry, we found that Cy. fabianii can be distinguished from other species. In antifungal susceptibility test, voriconazole and echinocandins exhibited good antifungal activities against the majority of Cy. fabianii isolates despite the absence of standard criteria.
1. Introduction
The incidence of yeast infections caused by non-albicans Candida (NAC) species has increased enormously. NAC species as well as Candida albicans can infect a broad range of body sites as opportunistic pathogens. With the increase in the number of patients showing acquired immune deficiency syndrome, the fungal infections caused by NAC species have been reported with a higher incidence rate [Citation1]. Cyberlindnera fabianii (basionym Hansenula fabianii), one of the NAC species, is known as a rare cause of invasive human infection. It has been recorded to cause pneumonia, endocarditis, prostatitis, fungemia and catheter-related infections [Citation2]. Recently, the case reports of fungal infections caused by Cy. fabianii in blood and urine have increased worldwide such as in China, Croatia, India, Czech and South Korea [Citation1,Citation3–5].
In recent years, the occurrence of Cy. fabianii has been reported in Korea due to growing awareness of its clinical significance. There have been three case reports of Cy. fabianii, which were all isolated from bloodstream [Citation4,Citation6,Citation7]. In addition, as part of a project of fungal pathogen culture collection in International St. Mary’s Hospital during 2016–2017, eleven isolates from eleven patients with different clinical histories were obtained. In this study, our objectives are to identify Cy. fabianii isolated from urine, blood and sputum, to characterize this species by using morphological and molecular methods and Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS), and to compare their Minimum Inhibitory Concentrations (MICs) against nine antifungal agents.
2. Materials and methods
2.1. Fungal isolates
During 2016–2017, yeast isolates were acquired from various clinical samples of the patients visiting International St. Mary’s Hospital like urine, blood, and sputum. The samples were directly inoculated on Sabouraud dextrose agar (SDA, Oxoid Ltd., Hampshire, UK) and incubated at 25 °C for 24 h. Although same species was isolated repeatedly, only one isolate from each patient was used in this study.
2.2. Morphological observations
The yeast cells incubated on the SDA were mounted in Shear’s mounting fluid on glass slides. An Olympus BX54 microscope equipped with an AxioCam MRc5 camera (Olympus, Tokyo, Japan) was used for measurements and imaging. The samples were examined in detail by means of bright-field and differential interference contrast light microscopy. We obtained 30 measurements for each structure at 400× and 1000× magnifications.
2.3. Molecular analysis
The yeast colonies were harvested from SDA and then genomic DNA was extracted using a DNeasy Plant Mini Kit (Qiagen Inc., Valencia, CA, USA). The internal transcribed spacer (ITS) region of rDNA and the D1/D2 region of large subunit (LSU) of rDNA were amplified using the primers ITS1 (5′-TCC GTA GGT GAA CCT GCG G-3′), ITS4 (5′-TCCTCC GCT TAT TGA TAT GC-3′) [Citation8], NL0R (5′-GCA TAT CAA TAA GCG GAG GAA AAG-3′) and LR5 (5′-CCG TGT TTC AAG ACG GG-3′) [Citation9]. The PCR parameters for the ITS and LSU amplifications were the following: an initial denaturation at 94 °C for 5 min, 35 cycles of denaturation at 95 °C for 1 min, annealing at 53 °C for 1 min, and extension at 72 °C for 2 min, and a final extension at 72 °C for 7 min. PCR amplicons were purified using a QIAquick PCR purification Kit (Qiagen Inc.) and then sequenced directly using Macrogen Sequencing Service (Microgen, Seoul, Korea).
2.4. Phylogenetic analyses
For phylogenetic analyses, the obtained sequences were assembled using the software SeqMan (Lasergene, DNASTAR, USA); in these analyses, ITS and LSU sequences were used. All available LSU sequences closely related to this species were retrieved from the NCBI GenBank database. The LSU sequence of Wickerhamomyces anomlalus was used as an outgroup taxon. Phylogenetic trees were generated by employing the neighbor-joining method with the maximum composite likelihood model by using MEGA6 [Citation10] with 1000 bootstrap replications.
2.5. Antifungal susceptibility testing
The antifungal susceptibility tests of Cy. fabianii isolates were conducted using Sensititre YeastOneTM YO10 methodology (Thermo Scientific, Cleveland, OH, USA). This procedure was conducted according to the manufacturer’s instructions. Cy. fabianii isolates were cultured on SDA and incubated at 35 °C for 24 h. Then, 20 µl of yeast suspension adjusted with 0.5 McFarland standard was transferred to 11 ml of YeastOne® inoculum broth. After that, 100 µl of the inoculated broth was transferred to the 96-well plates provided by the manufacturer. The plates were incubated at 35 °C for 24 h and the MIC endpoints were read. In total, nine antifungal agents were tested including amphotericin B, itraconazole, voriconazole, posaconazole, fluconazole, flucytosine, anidulafungin, micafungin, and caspofungin. Candida parapsilosis ATCC22019 was used for quality control.
2.6. MALDI-TOF MS
A single yeast colony from the SDA was transferred to a 1.5 ml tube. Then, 0.3 ml of water was added to suspend the colony and 0.7 ml of ethanol was further added. After 5 min centrifugation at 13,000 rpm, the supernatant was discarded and cell pellet was dried. Fifty microliters of 70% formic acid was added to the pellet and mixed. Fifty microliters of acetonitrile was added and mixed by vortex. After 5 min centrifugation at 13,000 rpm, 1.5 µl of supernatant was loaded onto a spot of MALDI plate and dried. 1.5 µl α-cyano-4-hydroxycinnamic acid (CHCA) solution was overlaid onto the spot of the dried sample. MALDI-TOF analysis for peptide profiling based identification was performed using MicroIDSys (ASTA, Suwon, Korea). Database of MicroIDSys (ver 1.23.2) contains 407 species (894 strains) of yeasts. The cut-off value of identification was set at ≥140 for all microorganisms.
3. Results
3.1. Medical history
During our 2016–2017 surveys, eleven isolates of Cy. fabianii were obtained from eleven patients with different medical histories in International St. Mary’s Hospital in Korea. Among them, eight isolates were obtained from urine, two isolates from sputum, and one isolate from blood. All patients had indwelling catheters including urine, chemoport and central venous catheter, and the ages ranged from 46 to 88. The medical histories of the patients are listed in .
Table 1. Medical history of the patients for the isolates used in this study.
3.2. Morphological characterization
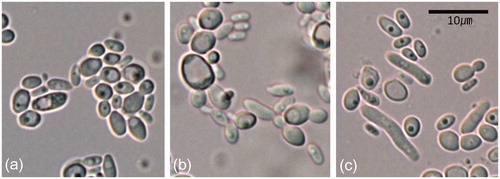
The yeast colonies were cream-colored, glistening and soft, usually smooth or partly wrinkled. Cells were unicellular, ovoid, ellipsoidal to broadly ellipsoidal, present in pairs or in groups, and 2.5 – 6.5 × 1.5 – 3.5 µm (. Conidiogenesis was a budding process and unipolar to bipolar (. Pseudohyphae was present (.
3.3. Phylogenetic analyses
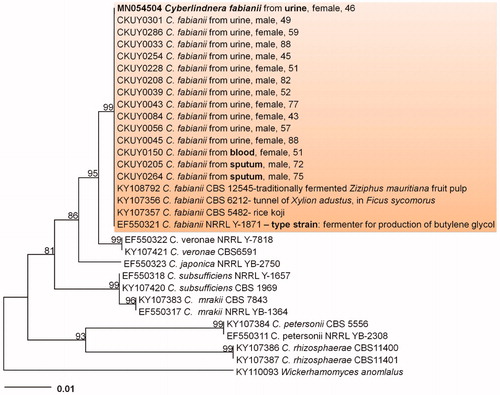
The resulting 583 bp ITS and 851 bp LSU sequences were obtained and deposited in GenBank (Accession Nos. MN054505 and MN054504). A GenBank BLAST search revealed that the ITS and LSU sequences showed 100% identity with those of Cy. fabianii (KY103044, KY107357). The phylogenetic analysis using LSU sequences revealed that the isolates in this study were placed in a distinct group in a neighbor-joining tree (). Thus, morphological characteristics and molecular data support to identify the isolates as Cy. fabianii.
3.4. Antifungal susceptibility testing
The MICs of each antifungal agent are shown in . There was no resistant isolate to amphotericin B, flucytosine, itraconazole, fluconazole, voriconazole, posaconazole, anidulafungin, micafungin, and caspofungin for in vitro antifungal susceptibility testing. Due to unavailability of interpretative criteria for rare yeasts such as Cyberlindnera spp., the MIC results were determined according to the CLSI interpretative breakpoints of other Candida spp. [Citation11]. Exceptionally, MFCCKHY00084 strain showed slight resistance to azole antifungal agents including itraconazole, fluconazole, voriconazole, and posaconazole. In addition, our data showed no correlation of Cy. fabianii infection and patient’s medical histories. Although the isolates of Cy. fabianii were obtained from different clinical samples such as urine, blood and sputum, there was no association between the isolation source and the antifungal susceptibility.
Table 2 Antifungal susceptibility testing of Cyberlindnera fabianii (n = 11) using Sensititre YeastOneTM YO10.
3.5. MALDI-TOF MS
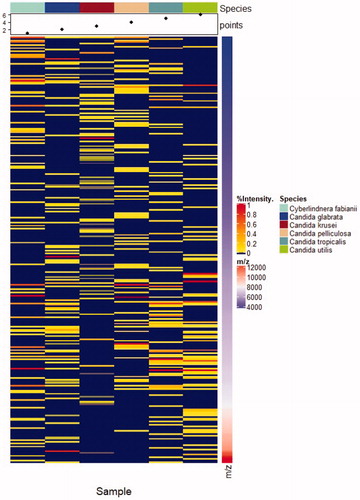
Cutoff score with typical 140 value was used for the identification with the ASTA MicroIDSys system. The results are presented in and . Among eleven isolates used in this study, eight isolates were confidently identified with acceptable score (over 140 for the ASTA MicroIDSys system). Although three isolates of Cy. fabianii were with an ASTA MicroIDSys score <140, two isolates were reliably identified as Cy. fabianii by MALDI-TOF MS with one isolate (MFCCKHY00208) identified as Clostridium sporogenes.
Figure 3. The comparison of MALDI-TOF profiles of Candida and its related species. The species used in the comparison were Cy. fabianii, C. glabrata, C. krusei, C. pelliculosa, C. tropicalis and C. utilis and MALDI-TOF profiles of each species were presented from left to right. MALDI-TOF profiles mainly between m/z 4000 and 12000 were compared. MicoIDSys MALDI-TOF system was used for this comparison.
4. Discussion
With the advent of advanced DNA sequencing technique, cryptic species and previously unrecognized species within species complex have been identified, along with changes in fungal taxonomy. The genus Cyberlindnera was established as a replacement name for the illegitimate name Lindnera [Citation12]. Minter [Citation12] introduced new generic name Cyberlindnera with twenty-one new combinations. Accordingly, Candida fabianii, Hansenula fabianii and Pichia fabianii were synonymized with Cy. fabianii. Although human infections caused by Cy. fabianii were known to be extremely rare [Citation13,Citation14], case reports of fungal infections caused by Cy. fabianii have been recently increasing worldwide including China, Croatia, India, USA and South Korea. Hamal et al. [Citation13] reported Pichia fabianii as the causative agent of endocarditis obtained from the blood of a patient. In addition, Wu et al. [Citation3] reported P. fabianii blood infection in a premature infant in China. In India, the first case of isolation of Cy. fabianii was from a urine sample of an immunocompromised patient [Citation15]. In South Korea, there have been three case reports of Cy. fabianii from bloodstream [Citation4,Citation6,Citation7]. In a recent survey of Czech hospitals, Cy. fabianii has emerged as a significant yeast with the same frequency as Candida lusitaniae and Candida guilliermondii [Citation1]. In addition, Hof et al. [Citation16] reported that neonates are relatively prone to the infection by Cy. fabianii.
The increased infections caused by NAC species have resulted in the need for an accurate identification of Candida spp., which is important for proper management of patients and prevention of emergence of drug resistance. According to our results of API 20 C and Vitek 2 using eleven isolates of Cy. fabianii, two isolates were identified as Candida utilis and the rests of them were Candida pelliculosa (), which was consistent with the study of Baghdadi et al. [Citation14]. That study revealed that Vitek 2 with ID 32 C system misidentified Cy. fabianii as C. pelliculosa or C. utilis. Bhally et al. [Citation17] also reported that API 20 C did not identify the yeast. In addition, Svobodova et al. [Citation1] mentioned that morphological identification and common biochemical kits cannot distinguish non-albicans clinical isolates such as C. pelliculosa, Cy. jadinii (previously C. utilis), and Cy. fabianii. It might be because there was no information of Cy. fabianii in any yeast database used in biochemical kit such as API 20 C AUX, ID 32 C, and Vitek-2 C [Citation1,Citation18]. Accordingly, only a few case reports of human infection caused by Cy. fabianii have been published. Thus, if molecular identification is performed or yeast database in biochemical kit is expanded, the number of case reports of Cy. fabianii is assumed to increase rapidly worldwide.
For reliable fungal species identification, DNA sequencing of ITS region and D1/D2 region of LSU gene is confirmed in many instances [Citation7,Citation15]. Thus, we performed molecular analysis with two markers ITS and LSU and showed good resolution to distinguish this species from other Cyberlindnera spp (). Although Gabriel et al. [Citation18] suggested that EF1-a gene is more effective than ITS region in order to identify accurately new emerging opportunistic yeast pathogens, this study showed that ITS sequence was enough to distinguish Cyberlindnera spp. Moreover, Valenza et al. [Citation19] already mentioned that ITS region allowed accurate identification of Cy. fabinaii. As the yeast species are difficult to identify based on physiological characteristics, the importance of molecular genetic-based assays has been emphasized in medical mycology [Citation20].
In the study of Mlinarić-Massoni et al. [Citation5], there was no resistance to amphotericin B, flucytosine, fluconazole, itraconazole, and voriconazole for in vitro antifungal susceptibility testing of Cy. fabianii. In addition, in vivo, Cy. fabianii from the blood or urine was clinically eliminated using fluconazole with liposomal amphotericin B or caspofungin. In this study, except one strain which showed slight resistance to azole antifungal susceptibility, most isolates of Cy. fabianii were susceptible to nine antifungal agents including amphotericin B, voriconazole, fluconazole, posaconazole, itraconazole, flucytosine, and echinocandins. Until now in Korea, Cy. fabianii isolates originated only from bloodstream and antifungal susceptibility testing was conducted on one isolate using anidulafungin [Citation6]. Thus, it is the first report to perform in vitro antifungal susceptibility test using nine antifungal agents in order to select appropriate and effective antifungal therapy and supervise the development of resistance.
Interest in MALDI-TOF MS is increasing as an alternative method for a reliable identification in the clinical setting [Citation21]. A number of studies have already been tried to evaluate the use of MALDI-TOF MS platforms for identifying clinical yeast isolates [Citation20]. Especially, MALDI-TOF MS is an accurate and reliable tool for identification of bloodstream yeasts [Citation22]. The comparison between our molecular data and MALDI-TOF data confirmed that MALDI-TOF using MicroIDSys (ASTA, Seoul, Korea) could be a reference method for accurate identification of Cyberlindnera spp. (). Although one isolate was misidentified in this study, ASTA platforms showed discriminating powers of the isolates of Cy. fabianii from other NAC. Moreover, through this study, database of MicroIDSys (ver 1.23.2) is expanding to include rare yeasts such as Cyberlindnera spp. and other NAC spp. The mass spectral profiles of Cy. fabianii would expand the current databases by performing MALDI-TOF continuously. In a recent case report, the correct identification of this species was proved by MALDI-TOF method using Bruker Daltonic (Bremen, Germany) [Citation16]. Therefore, MALDI-TOF MS will be soon a promising tool for rapid yeast diagnostics and outbreak investigation.
The isolates of Cy. fabianii have been obtained from various clinical samples including blood, stool, urine, nasopharyngeal swab, ureostomal swab, wound, gasric content, tubal aspirate, and oropharyngeal swab [Citation5]. In this study, the most frequent isolated samples were the urine, followed by sputum and blood, which was a reminiscent of the use of indwelling catheters such as urinary catheter, central catheter and chemoport. Although Valenza et al. [Citation19] did not describe a causal relationship between the disease and the presence of Cy. fabianii clearly, it was thought that this infection was clearly associated with indwelling catheter use, especially urinary catheter. Urinary tract infection caused by use of an indwelling urinary catheter is the most common nosocomial infection in patients [Citation23,Citation24]. Of note, all patients infected by Cy. fabianii in this study were submitted to multiple invasive medical procedures during their hospitalization with central venous catheter, surgery and antibacterial therapy. Catheter-related blood infections caused by Cy. fabianii have already been reported in several case studies [Citation4,Citation14]. Kaur et al. [Citation25] also mentioned that the frequent use of central venous catheters, urinary catheters, and prosthetic devices lead to a high risk infection of Candida species. As a result of comparison between patient’s medical histories and isolated samples in this study, urinary infection caused by Cy. fabianii occurred in patients with the urine foley catheter and a patient with cervical vein catheter is prone to be bloodstream infection.
Most isolates were obtained from urine in this study, indicating that this species could be frequently found in the urine. Yeasts, especially Candida spp., detected in urine are called candiduria [Citation26]. They often occur in patients who have bladder colonization, and in patients who have upper urinary tract infection [Citation27]. Most cases of candiduria are asymptomatic and there is no need of antifungal therapy in asymptomatic candiduria [Citation26]. According to the surveillance study of candiduria, there were common risk factors such as urinary tract instrumentation, prior surgical procedures, recent use of antibiotics, advanced age, female sex, diabetes mellitus, immunosuppressive therapy and prolonged hospital stay. The rate of acquisition of candiduria is known to be tightly linked to women and older persons, which is consistent with our results. As there was no symptoms in the patients infected by Cy. fabianii, urinary track candidiasis was confirmed throughout direct and culture examinations of urine sample. Thus, further study of relationship between Cyberlindnera spp. and urinary tract infection should be performed in future.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Svobodova L, Bednarova D, Ruzicka F, et al. High frequency of Candida fabianii among clinical isolates biochemically identified as Candida pelliculosa and Candida utilis. Mycoses. 2016;59:241–246.
- Grenouillet F, Millon L, Chamouine A, et al. Pichia fabianii fungemia in a neonate. Pediatr Infect Dis J. 2010;29:191.
- Wu Y, Wang J, Li W, et al. Pichia fabianii blood infection in a premature infant in China: case report. BMC Res Notes. 2013;6:77.
- Yun JW, Park KS, Ki CS, et al. Catheter-related bloodstream infection by Lindnera fabianii in a neutropenic patient. J Med Microbiol. 2013;62:922–925.
- Mlinarić-Missoni E, Hatvani L, Kocsubé S, et al. Cyberlindnera fabianii in the neonatal and paediatric intensive care unit: case reports. JMM Case Rep. 2015;2:1–6. https://doi.org/10.1099/jmmcr.0.000032.
- Lee JI, Yu S, Park JS, et al. Successful treatment of fungemia caused by Cyberlindnera fabianii with Anidulafungin: a case report. Ann Clin Microbiol. 2015;18:94–97.
- Kim YJ, Yang JJ, Lee HJ. Possibility of frequent detection of invasive Cyberlindnera fabianii infection using molecular method. Ann Clin Microbiol. 2015;18:133–134.
- White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego, CA, USA: Academic Press; 1990. p. 315–322.
- O'Donnell K. Fusarium and its near relatives In: Reynolds DR, Taylor JW, editors. The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. Wallingford, United Kingdom: CAB International; 1993. p. 529–580.
- Tamura K, Stecher G, Peterson D, et al. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729.
- Clinical and Laboratory Standards Institute (CLSI). M27-S4 Reference method for broth dilution antifungal susceptibility testing of yeasts: fourth informational supplement M27-S4. Wayne, PA: Clinical and Laboratory Standards Institute; 2012.
- Minter DW. Cyberlindnera, a replacement name for Lindnera kurtzman et al., nom. illegit. Mycotaxon. 2009;110:473–476.
- Hamal P, Ostransky J, Dendis M, et al. A case of endocarditis caused by the yeast Pichia fabianii with biofilm production and developed in vitro resistance to azoles in the course of antifungal treatment. Med Mycol. 2008;46:601–605.
- Baghdadi J, Hemarajata P, Humphries R, et al. First report of ventriculoperitoneal shunt infection due to Cyberlindnera fabianii. Case Rep Infect Dis. 2015;630816:1–6.
- Jindal N, Arora S, Dhuria N, et al. Cyberlindnera (Pichia) fabianii infection in a neutropenic child: importance of molecular identification. JMM Case Rep. 2015;2:1–3. http://dx.doi.org/10.1099/jmmcr.0.000033.
- Hof H, Amann V, Tauber C, et al. Peritonitis in a neonate due to Cyberlindnera fabianii, an ascomycetic yeast. Infection. 2017;45:921–924.
- Bhally HS, Jain S, Shields C, et al. Infection in a neonate caused by Pichia fabianii: importance of molecular identification. Med Mycol. 2006;44:185–187.
- Gabriel F, Noel T, Accoceberry I. Lindnera (Pichia) fabianii blood infection after mesenteric ischemia. Med Mycol. 2012;50:310–314.
- Valenza G, Valenza R, Brederlau J, et al. Identification of Candida fabianii as a causal of lethal septicaemia. Mycoses. 2006;49:331–334.
- Zhao Y, Tsang CC, Xiao M, et al. Yeast identification by sequencing, biochemical kits, MALDI-TOF MS and rep-PCR DNA fingerprinting. Med Mycol. 2018;56:816–827.
- Haas M, Grenouillet F, Loubersac S, et al. Identification of cryptic Candida species by MALDI-TOF mass spectrometry, not all MALDI-TOF systems are the same: focus on the C. parapsilosis species complex. Diagn Microbial Infect Dis. 2016;84:385–386.
- Ghosh AK, Paul S, Sood P, et al. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry for the rapid identification of yeasts causing bloodstream infections. Clin Microbiol Infect. 2015;21:372–378.
- Richard MJ, Edwards JR, Culver DH, et al. Nosocomial infections in medical intensive care units in the United States. National Nosocomial Infections Surveillance System. Crit Care Med. 1999;27:887–892.
- Nicolle LE. Catheter associated urinary tract infections. Antimicrob Resist Infect Control. 2014;3:23.
- Kaur R, Dhakad MS, Goyal R, et al. Emergence of non-albicans Candida species and antifungal resistance in intensive care unit patients. Asian Pac J Trop Biomed. 2016;6:455–460.
- Bukhary ZA. Candiduria: a review of clinical significance and management. Saudi J Kidney Dis Transpl. 2008;19:350–360.
- Kauffman CA. Candiduria. Clin Infect Dis. 2005;41:S371–S376.