Abstract
During an investigation of fungi of the order Mucorales from freshwater and sediment samples in Korea, we isolated six strains, NNIBRFG6649, NNIBRFG6255, NNIBRFG1498, CNUFC-YJ13, CNUFC-YR7, and NNIBRFG2739. The morphology and phylogeny of these strains were analyzed. Based on the morphological characteristics and molecular data from internal transcribed spacer (ITS) region, the isolates NNIBRFG6649 and NNIBRFG6255 were identified as Mucor abundans, and M. aligarensis, respectively. The isolates NNIBRFG1498 and CNUFC-YJ13 were identified as M. moelleri, whereas the isolates CNUFC-YR7 and NNIBRFG2739 were identified as M. heterogamus. To the best of our knowledge, M. abundans, M. aligarensis, M. moelleri, and M. heterogamus have not yet been reported in Korea.
Keywords:
1. Introduction
The genus Mucor was described by Fresenius in 1850 [Citation1]; it belongs to the family Mucoraceae, order Mucorales, subphylum Mucoromycotina, and phylum Mucoromycota, and comprises the largest number of species within Mucorales [Citation2,Citation3]. Mucor species are characterized by the formation of non-apophysate sporangia on simple or branched sporangiophores, and by zygospores that are borne on opposed [Citation4] or tong-like suspensors [Citation5].
More than 50 species have been reported in the genus Mucor by applying a polyphasic approach using molecular and morphological characters [Citation6]. However, this list is rapidly increasing with many new taxa of Mucor being described worldwide recently [Citation7–10]. Mucor species are commonly isolated from soil, fruit, vegetables, stored grains, insects, or dung [Citation5,Citation11].
Species belonging to this genus are valued in the biotechnology industry due to their ability to produce various secondary metabolites [Citation12,Citation13]. Mucor species are used in biotechnological processes such as bioremediation [Citation14–16]. In addition, some Mucor species have been used in the fermentation of traditional Asian foods [Citation17], whereas others are considered to be the causal agents of cutaneous zygomycosis in humans [Citation18].
Mucor species have been identified traditionally by their morphological characteristics such as the size and shape of their sporangia, and their mode of reproduction. Recently, molecular data have revealed the phylogenetic position of Mucor and the relationship among its species [Citation5]. Based on the phylogeny of ITS and large subunit rDNA regions of several mucoralean species, Walther et al. [Citation5] observed that some Mucor species with transitory curved sporangiophores were grouped with Backusella Hesselt. & J.J. Ellis. Therefore, these Mucor species were transferred to the genus Backusella. In Korea, ten Mucor species have been reported as new records and four as new species [Citation7,Citation8,Citation19,Citation20].
During an inventory of fungal species from freshwater and sediment samples, four Mucor species were identified. The objective of the present study was to perform morphological and molecular characterization of the undescribed Mucor species in Korea: M. abundans, M. aligarensis, M. moelleri, and M. heterogamus.
2. Materials and methods
2.1. Isolation of fungal strains from freshwater and sediment samples
Freshwater and sediment samples were collected from different geographical regions of the Republic of Korea. Sampling information for the present study is shown in . The samples were transported in sterile 50-mL tubes and were stored at 4 °C until required. The media and method used in this study were based on the protocols described by Nguyen et al. [Citation20] and Mun et al. [Citation21]. The plates were incubated at 20 °C and 25 °C for 2–5 d. Pure isolates were obtained by picking individual colonies of varied morphologies, transferring them onto potato dextrose agar (PDA; 39 g of potato dextrose agar in 1 L of deionized water) plates, and sub-culturing them until pure mycelia were obtained. For stock storage, pure isolates were maintained in PDA slant tubes with 20% glycerol at −80 °C at the Environmental Microbiology Laboratory Fungarium, Chonnam National University, Gwangju, Korea, and were also deposited at the Culture Collection of the Nakdonggang National Institute of Biological Resources (NNIBR, Sangju, Korea).
Table 1. Information about sampling and all the isolates used in the present study.
2.2. Morphological studies
Pure cultures of Mucor spp. were cultured in triplicate on PDA, malt extract agar (MEA; malt extract 2%; agar 2% in 1 L of deionized water), oatmeal agar (OA; Difco, Sparks, MD), Czapeck-dox solution agar (CDA; Difco), corn meal agar (CMA; Difco), potato carrot agar (PCA; HiMedia, Mumbai, India), Dichloran glycerol chloramphenicol (DG18; Merck Millipore, Billerica, MA), and yeast extract peptone dextrose agar (YPDA; Duchefa Biochemie, Haarlem, Netherlands) media and incubated at 5, 15, 20, 25, 30, 35, and 40 °C. Colony growth was monitored for 7 d and measured every 24 h. Fragments of mycelia were removed from the cultures, placed on microscope slides with lactic acid (60%). The Olympus BX51 microscope with differential interference contrast optics (Olympus, Tokyo, Japan) was used to obtain digital images.
2.3. DNA extraction, polymerase chain reaction, and sequencing
Genomic DNA was extracted directly from the mycelia of fungal isolates using a SolGent Genomic DNA Prep Kit (SolGent Co. Ltd., Daejeon, Korea). The internal transcribed spacer (ITS) region was amplified using the primer pairs ITS1 and ITS4 [Citation22]. The PCR reaction mixture (total volume, 20 µL) contained a fungal DNA template, 5 pmol/µL of each primer, AccuPower PCR PreMix (Bioneer Corp., Daejeon, Korea). The PCR products were purified using an AccuPrep PCR Purification Kit (Bioneer Corp.) according to the manufacturer’s instructions. Sequencing was performed using the same PCR primers, and run on an ABI PRISM 3730XL genetic analyzer (Applied Biosystems, Waltham, MA).
2.4. Phylogenetic analysis
The fungal sequences obtained from GenBank database () were aligned using MAFFT [Citation23] and then confirmed in MEGA6 [Citation24]. Phylogenetic analyses were performed using MEGA6 software [Citation24], and a maximum-likelihood (ML) tree was constructed using Kimura’s two-parameter correction method. Umbelopsis isabellina was used as an outgroup. The sequences of NNIBRFG6649, NNIBRFG6255, NNIBRFG1498, CNUFC-YJ13, CNUFC-YR7, and NNIBRFG2739 were deposited at the National Center for Biotechnology Information (NCBI) database under the accession numbers as shown in .
Table 2. Taxa, collection numbers, and GenBank accession numbers used in the present study.
3. Results
3.1. Phylogenetic analysis
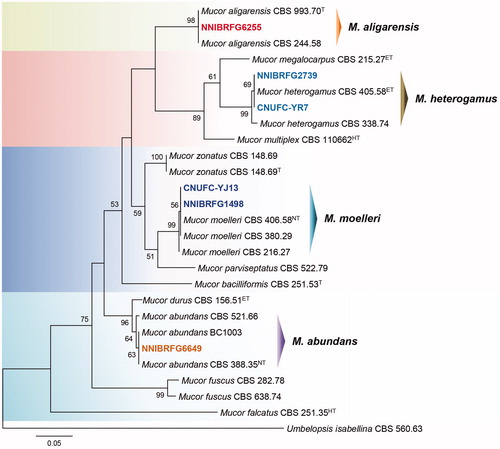
ITS rDNA data were used to determine the phylogenetic relationships between NNIBRFG6649, NNIBRFG6255, NNIBRFG1498, CNUFC-YJ13, CNUFC-YR7, and NNIBRFG2739, and related species. A BLASTn search of ITS rDNA revealed that the isolate NNIBRFG6649 matched M. abundans (GenBank accession no. MH971277) with a similarity value of 100% (581/581 bp); isolate NNIBRFG6255 matched M. aligarensis (GenBank accession no. MH857771) with 100% similarity (571/571 bp); isolates NNIBRFG1498 and CNUFC-YJ13 matched M. moelleri (GenBank accession no. MH857827) with 100% similarity (492/492 bp); and isolates CNUFC-YR7 and NNIBRFG2739 matched M. heterogamus (GenBank accession no. JN206167) with 99.6% similarity (518/520 bp). Phylogenetic analyses of the ITS sequences suggested that strains NNIBRFG6649, NNIBRFG6255, NNIBRFG1498, CNUFC-YJ13, CNUFC-YR7, and NNIBRFG2739 were placed within the same clade with M. abundans, M. aligarensis, M. moelleri, and M. heterogamus ().
Figure 1. Phylogenetic tree of M. abundans NNIBRFG6649, M. aligarensis NNIBRFG6255, M. moelleri CNUFC-YJ13 and NNIBRFG1498, and M. heterogamus CNUFC-YR7 and NNIBRFG2739, and related species based on a maximum-likelihood analysis of ITS rDNA sequences. The sequence of Umbelopsis isabellina was used as an outgroup. Numbers at the nodes indicate the bootstrap values (>50%) from 1000 replications. The bar indicates the number of substitutions per position. The new isolates from the present study are shown in bold.
3.2. Taxonomy
3.2.1. Taxonomy of NNIBRFG6649
Mucor abundans Povah, Bulletin of the Torrey Botanical Club 44: 292 (1917) ().
Figure 2. Morphology of Mucor abundans NNIBRFG6649. (A) Colony on potato dextrose agar; (B–D) Branched sporangiophores and sporangia; (E–G) Columella with clear collar present; (H) Sporangiospores. Scale bars: B–H = 20 μm.
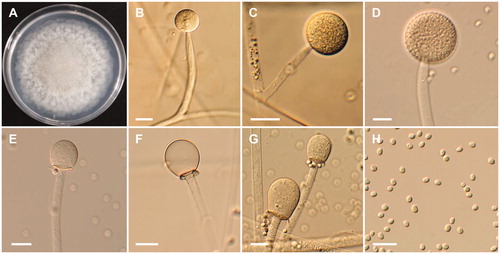
Description: The strain grew rapidly at 25 °C on PDA, and reached 80–82 mm in diameter after 5 d of incubation. The colony was initially white, but subsequently turned brown. The sporangiophores were 163.9–548.6 µm in length and 7.3–17.7 µm in width; they arose from aerial and submerged mycelia, and produced a large number of sporangia. The sporangia were globose to subglobose, 31–76.5 × 30.5–75 µm. Columellae with collars were subglobose to pyriform, 27.0–45.5 × 24.5–35.5 µm. Sporangiospores were hyaline, subglobose to short elliptical, and measured 4–5.8 × 3.4–4.5 µm. Chlamydospores were present. Zygosporangia were not observed.
3.2.2. Taxonomy of NNIBRFG6255
Mucor aligarensis B.S. Mehrotra & B.R. Mehrotra, Sydowia 23 (1–6): 183 (1970)
=Mucor petrinsularis var. ovalisporus G. Sm., Transactions of the British Mycological Society 40: 482 (1957) ().
Figure 3. Morphology of Mucor aligarensis NNIBRFG6255. (A) Colony on potato dextrose agar; (B–D) Sporangiophores and sporangia; (E–G) Columella with clear collar present; (H) Sporangiospores. Scale bars: B–H = 20 μm.
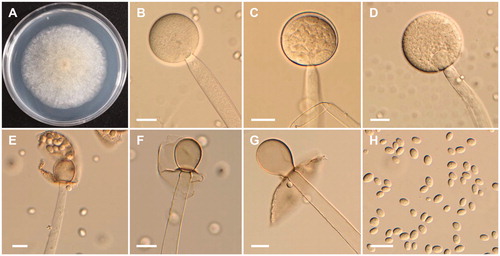
Description: The strain grew rapidly at 20 °C on PDA, and reached 67–70 mm in diameter after 5 d of incubation. The colony was initially white, but subsequently turned brown. The sporangiophores were 957.7–1056.4 µm in length and 8.4–21 µm in width; they arose from aerial and submerged mycelia, septate and produced a large number of sporangia. The sporangia were globose to subglobose, 26.5–102.6 × 23.0–102 µm, and were borne terminally on the main axes or branches of the sporangiophores. Columellae with large collars were subglobose, obovoid to pyriform, 23.5–55 × 18.5–41.5 µm. Sporangiospores were hyaline, subglobose to oval, and measured 7.5–10 × 5.5–8.5 µm. Zygosporangia were not observed.
3.2.3. Taxonomy of CNUFC-YJ13
Mucor moelleri (Vuill.) Lendn., Matériaux pour la Flore Cryptogamique Suisse 3 (1): 72 (1908)
≡Zygorhynchus moelleri Vuill., Bulletin de la Société Mycologique de France 19: 117 (1903) ().
Figure 4. Morphology of Mucor moelleri CNUFC-YJ13. (A) Colony on potato dextrose agar; (B–D) Branched sporangiophores and sporangia; (E) Columella with clear collar present; (F) Sporangiospores; (G, H) Zygospores. Scale bars: B–H = 20 μm.
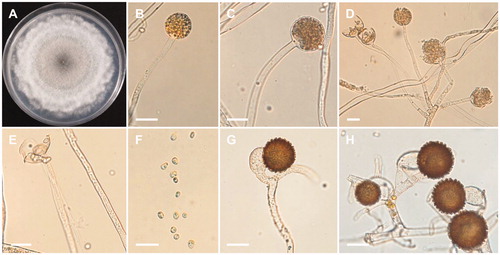
Description: The strain grew rapidly at 25 °C on PDA, spreading completely within the petri dish (90 mm in diameter) after 5 d of incubation. The initial color of the colony was white, which later turned gray. Sporangiophores arose from aerial and submerged mycelia, the formation of numerous zygosporangia was observed. Sporangia were globose to subglobose, and measured 25.6–48.8 × 24.6–42.3 µm, and were borne terminally on the main axes or branches of sporangiophores. Columellae were conical to globose. Sporangiospores were subglobose, oval, ellipsoidal, and measured 4.3–8.5 × 3.9–6.8 µm. Zygospores were globose to subglobose, and measured 17.3–41.3 × 14.3–40.6 µm, reddish-brown when young, and subsequently turned black. The strain was homothallic.
3.2.4. Taxonomy of CNUFC-YR7
Mucor heterogamus Vuill., Bull. Séanc. Soc. Sci. Nancy: 8: 50 (1887)
≡Zygorhynchus heterogamus (Vuill.) Vuill., Bulletin de la Société Mycologique de France 19: 117 (1903) ().
Figure 5. Morphology of Mucor heterogamus CNUFC-YR7. (A) Colony on potato dextrose agar; (B–D) Branched sporangiophores and sporangia; (E–G) Zygospores. Scale bars: B–G = 20 μm.
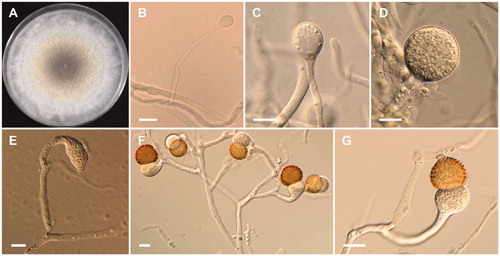
Description: The strain grew rapidly at 25 °C on PDA, and completely filled the petri dish (90 mm in diameter) after 5 d of incubation. The colony was initially white, but subsequently turned pale yellow, with dark gray in the center. The sporangiophores were 3.5–15 µm in width and variable in length; they arose from aerial and submerged mycelia, and produced a small number of sporangia. The sporangia were globose to subglobose, 18.5–47.5 × 20.3–50 µm, and were borne terminally on the main axes or branches of the sporangiophores. The zygospores were globose, up to 66.5 µm in diameter, and were formed throughout the colony, but were most abundant just above the substrate; they were reddish-brown when young, and subsequently turned black. The suspensors were globose, pyriform, and unequal. The strain was homothallic.
4. Discussion
To date, only 14 species of Mucor have been reported in Korea [Citation7,Citation8,Citation19,Citation20]. Therefore, any data pertaining to this genus would increase our understanding of the diversity and geographical distribution of fungal species in Korea.
Here, we discuss the phylogeny and morphological characteristics of M. abundans, M. aligarensis, M. moelleri, and M. heterogamus. Walther et al. [Citation5] proposed the use of the ITS region as a standardized marker for the sequence analysis of Mucor species. The phylogenetic tree using ITS region showed that strains NNIBRFG6649, NNIBRFG6255, NNIBRFG1498 and CNUFC-YJ13, and CNUFC-YR7 and NNIBRFG2739 were grouped with the species M. abundans, M. aligarensis, M. moelleri, and M. heterogamus, respectively ().
The morphological characteristics of the M. abundans isolate studied were generally similar to those previously described by Povah [Citation25]. However, differences were observed in the size of sporangia from M. abundans with respect to that from the previous description. Our observations showed that the sporangia in isolate NNIBRFG6649 were smaller than those previously described by Povah [Citation25] (67–78 µm). Previously M. abundans was reported to be isolated from horse dung, sandy tilled soil, decaying tomato, rabbit dung, dog dung, and forest soil [Citation5,Citation25]. To our knowledge, this is the first report of M. abundans isolation from freshwater.
The isolate NNIBRFG6255 is morphologically similar to M. aligarensis with respect to the production of globose multispored sporangia, and subglobose to oval sporangiospores. However, sporangiosphore sizes reported in literature (6.6–15.4 × 5.5–11 µm) [Citation26] were bigger than (7.5–10 × 5.5–8.5 µm) those of our isolate. Isolate NNIBRFG6255 produces pyriform columellae, whereas pyriform columellae are absent in M. aligarensis described by Mehrotra and Mehrotra [Citation26]. M. aligarensis has been isolated from garden soil and the human ear [Citation5,Citation26]. This is first report of M. aligarensis isolation from freshwater.
The morphological characteristics of the isolate CNUFC-YJ13 in this study were similar to those previously described by Zheng and Liu [Citation27], as producing globose, multispored sporangia, conical to globose columellae. However, sporangia and zygospore sizes reported in the literature [Citation27] were bigger than those of our isolate. Sporangiospores in isolate CNUFC-YJ13 contains one or two droplets, which was not described by Zheng and Liu [Citation27]. CNUFC-YJ13 was confirmed as M. moelleri based on morphological observations and ITS phylogenetic comparison.
Although the morphological features of isolate CNUFC-YR7 were similar to those of M. heterogamus described by Hesseltine et al. [Citation28], there were differences in the diameters of the sporangia and zygospores. The sizes of sporangia reported in literature ranged from 15–55 µm [Citation28], which are slightly larger than our maximum measurements. The zygospores (30–70) µm [Citation28] were larger than those in our isolates. The molecular data confirmed the morphological identification of CNUFC-YR7 as M. heterogamus.
Several species of Mucor are of great interest to the biotechnology industry because they can produce proteolytic enzymes and secondary metabolites such as phytoalexin elicitor [Citation13,Citation29]. Somkuti [Citation30] showed that M. pusillus and M. miehei produce cellulase enzymes. Andrade et al. [Citation31] demonstrated high protease production by M. circinelloides using glucose as a substrate. Future studies should investigate the abilities of Mucor species to produce extracellular enzymes, and their potential use in biotechnology.
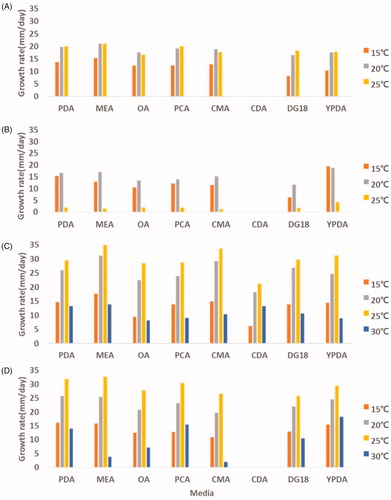
In general, growth temperature plays an important role in determining the physiological characteristics of fungi. Especially, Mucor spp. are able to cause infections in humans. These isolates grow over a wide range of temperatures with varying growth rates on PDA, MEA, OA, PCA, CMA, CDA, DG18, and YPDA (). The optimal growth temperature range is 20–25 °C, except for M. aligarensis (15–20 °C). Among the four species, the minimal and maximal temperatures for which Mucor growth was observed were below 15 °C (M. abundans, M. aligarensis, M. moelleri, and M. heterogamus) and 30 °C (M. moelleri and M. heterogamus). M. aligarensis does not grow well at temperatures above 25 °C. MEA, PDA, and YPDA were favorable for growing all the isolates. The best mycelial growth was observed at 25 °C on MEA for M. abundans, M. moelleri, and M. heterogamus, whereas M. aligarensis grew well on YPDA at 15 °C. On all media, the isolates did not grow at 35 °C. Zygospores were not observed in the all media for M. abundans and M. aligarensis.
Figure 6. Effect of temperature and culture medium on mycelial growth of M. abundans (A); M. aligarensis (B); M. moelleri (C); M. heterogamus (D). Mycelia were grown on PDA, MEA, OA, PCA, CMA, CDA, DG18, and YPDA, at different temperatures, as indicated.
Fungi play a major role in the decomposition of complex organic compounds in freshwater ecosystems, thereby providing nutrients for other aquatic organisms. In addition, freshwater-derived fungi have been widely studied for their bioactive metabolites. Therefore, further studies are required for a better understanding of the biological role of secondary metabolites, such as extracellular enzyme activity, antibacterial, antifungal, and anticancer activities in fungi, especially in M. abundans, M. aligarensis, M. moelleri, and M. heterogamus.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Fresenius G. Beiträge zur Mykologie. Frankfurt: Hefte 1; 1850.
- Benny GL, Humber RA, Voigt K. Zygomycetous fungi: Phylum Entomophthoromycota and subphyla Kickxellomycotina, Mortierellomycotina, Mucoromycotina, and Zoopagomycotina. In: McLaughlin DJ, Spatafora JW, editors. Mycota VII, Part A, Systematics and evolution. New York (NY): Springer-Verlag; 2014. p. 209–250.
- Spatafora JW, Benny GL, Lazarus K, et al. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia. 2016;108(5):1028–1046.
- Schipper M. On certain species of Mucor with a key to all accepted species. Stud Mycol. 1978;17:1–52.
- Walther G, Pawłowska J, Alastruey-Izquierdo A, et al. DNA barcoding in Mucorales: an inventory of biodiversity. Persoonia. 2013;30(1):11–47.
- Gherbawy Y, Kesselboth C, Elhariry H, et al. Molecular barcoding of microscopic fungi with emphasis on the Mucoralean genera Mucor and Rhizopus. In: Gherbawy Y, Voigt K, editors. Molecular identification of fungi. Berlin, Heidelberg: Springer; 2010. p. 213–253.
- Wanasinghe DN, Phukhamsakda C, Hyde KD, et al. Fungal diversity notes 709–839: taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers. 2018;89(1):1–236.
- Phookamsak R, Hyde KD, Jeewon R, et al. Fungal diversity notes 929–1035: taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Divers. 2019;95(1):1–273.
- De Lima CLF, Lima DX, De Souza CAF, et al. Description of Mucor pernambucoensis (Mucorales, mucoromycota), a new species isolated from the Brazilian upland rainforest. Phytotaxa. 2018;350(3):274.
- De Souza CAF, Voigt K, Gurgel LS, et al. A new species of Mucor (Mucoromycotina, Mucorales) isolated from an enclave of Upland Atlantic Forest in the semi-arid region of Brazil. Phytotaxa. 2018;351(1):53.
- Benny GL. The methods used by Dr. R. K. Benjamin, and other mycologists, to isolate Zygomycetes. Aliso. 2008;26(1):37–61.
- Kosa G, Zimmermann B, Kohler A. High-throughput screening of Mucoromycota fungi for production of low- and high-value lipids. Biotechnol Biofuels. 2018;11:66.
- de Souza PM, Bittencourt MLA, Caprara CC, et al. A biotechnology perspective of fungal proteases. Braz J Microbiol. 2015;46(2):337–346.
- Purnomo AS, Koyama F, Mori T, et al. DDT degradation potential of cattle manure compost. Chemosphere. 2010;80(6):619–624.
- Srinivasan A, Viraraghavan T. Oil removal from water using biomaterials. Bioresour Technol. 2010;101(17):6594–6600.
- Jabasingh SA, Pavithra G. Response surface approach for the biosorption of Cr6+ ions by Mucor racemosus. CLEAN Soil Air Water. 2010;38(5-6):492–499.
- Nout MJR, Asian AK. Fungal fermented food. In: Hofrichter M, editor. Industrial applications. Berlin: Springer; 2010. p. 29–58.
- Ribes JA, Vanover-Sams CL, Baker DJ. Zygomycetes in human disease. Clin Microbiol Rev. 2000;13(2):236–301.
- Lee YS, Jung HY, Lee HB, et al. KSM. National list of species of Korea Ascomycota, Glomeromycota, Zygomycota, Myxomycota, Oomycota. National Institute of Biological Resources Ministry of Environment, Korea. 2015.
- Nguyen TTT, Lee HB. Isolation and characterization of three Zygomycetous fungi in Korea: Backusella circina, Circinella muscae, and Mucor ramosissimus. Mycobiology. 2018;46(4):317–327.
- Mun HY, Goh J, Oh Y, et al. First report of three Didymella species isolated from freshwater ecosystem in Korea. Kor J Mycol. 2018;46(2):91–97.
- White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols. London: Academic Press; 1990. p. 315–322.
- Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2017;20(4):1160–1166.
- Tamura K, Stecher G, Peterson D, et al. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729.
- Povah A. A critical study of certain species of Mucor. Bull Torrey Bot Club. 1917;44(6):287–310.
- Mehrotra BS, Mehrotra BR. Two new species of Mucor from India – IV. Sydowia. 1969;23:183–185.
- Zheng RY, Liu XY. More taxa of Zygorhynchus from China. Sydowia. 2010;59:273–372.
- Hesseltine CW, Benjamin CR, Mehrotra BS. The genus Zygorhynchus. Mycologia. 1959;51(2):173–194.
- Simões K, Dietrich SMC, Hahn MG, et al. Purification and characterization of a phytoalexin elicitor from spores of the saprobe Mucor ramosissimus. Rev Bras Bot. 2005;28(4):735–744.
- Somkuti GA. Synthesis of cellulase by Mucor pusillus and Mucor miehei. J Gen Microbiol. 1974;81(1):1–6.
- Andrade VS, Sarubbo LA, Fukushima K, et al. Production of extracellular proteases by Mucor circinelloides using D-glucose as carbon source/substrate. Braz J Microbiol. 2002;33:106–110.
