Abstract
Fungal contamination of built-in furniture is a frequent problem in Korea when new apartment is built. However, domestic information on the contaminating fungi is very limited. This study was conducted to isolate, identify and characterize the fungi of the problem in one of the apartment houses where the fungi were claimed in the built-in furniture before the house owner moves in. Fungi present in the furniture installed in a main room, dress room, and kitchen side were visually and microscopically confirmed and purely isolated on PDA. The isolated fungi were identified by analyzing the morphological characteristics and nucleotide sequence of the ITS, calmodulin gene, and TEF-1α gene. Aspergillus creber, A. niger, A. pseudoglacus, A. ruber, Cladosporium perangustum and Penicillium commune were identified. Four out of the six fungal species were positive for at least one enzyme in six kinds of extracellular enzyme assays. When these four species (A. creber, A. niger, C. perangustum and P. commune) were inoculated onto four kinds of wood chips of furniture materials, they were able to colonize all of the wood chips. Their settlement was better at 95% humidity condition than at 30% humidity condition. Among the four species, C. perangustum caused the darkest discoloration and secreted the most number of extracellular enzymes. The four species were re-isolated from the colonized wood chips and confirmed as the problematic fungi in the built-in furniture.
1. Introduction
With the progress of urbanization, the construction of apartment and high-rise buildings has been increased across countries in Korea. The increase in the number of such housing construction has made the installation of built-in furniture much more commonly due to the convenience that can be used immediately after moving. Amid these trends, the emission of various pollutants has been increasing as the use of various building materials and household goods has increased. Examples of such indoor air pollutants are nitrogen dioxide, carbon dioxide, formaldehyde, radon, ozone, volatile organic compounds and microorganisms [Citation1].
Regarding formaldehyde, it is a valuable precursor to diverse materials and chemical compounds. When treated with urea or melamine, formaldehyde produces urea formaldehyde resin and melamine resin. These resins are used as adhesives for manufacturing of hardwood plywood, particle board and medium density fiberboard [Citation2]. Formaldehyde resin also has the effect of preservation from fungal decay. Since hardwood plywood, particle board and medium density fiberboard are construction materials for wooden buildings and furniture in homes, it is one of common indoor air pollutants originated from many homes [Citation3]. Since we spend more than 60–80% of our time living in a house, health concern with such indoor air pollutants is manifest [Citation1]. At concentrations above 0.1 ppm in air, formaldehyde can irritate our eyes and mucous membranes, resulting in watery eyes [Citation4]. Formaldehyde inhaled at this concentration may cause headaches, a burning sensation in the throat, and difficulty breathing, and can trigger or aggravate asthma symptoms [Citation5,Citation6]. Thus, the Ministry of Environment of Korean Government has been making efforts to reduce the use of volatile organic compounds such as toluene and formaldehyde through legislation of the Indoor Air Quality Management Law [Citation7].
In 2018, fungi were added to the list of biological pollutants that needs to be regulated under the Indoor Air Quality Management Law [Citation7]. Thus, fungi are now officially recognized as health concerned organisms in Korea. According to the reports of diversity in indoor and outdoor air quality, Cladosporium, Penicillium, Aspergillus and Alternaria are the dominant species [Citation8]. Penicillium brevicompactum, Aspergillus versicolor and Stachybotrys chartarum have been reported to produce fungal toxins [Citation9]. Actual case studies of fungi are lacking in Korea. When it comes to residential space for Koreans, apartments are gaining popularity as a major residential space, and in which furniture has recently been popularly installed in the form of built-in furniture. To the pace with this trend of built-in furniture installation, there have been problems with fungal contamination in the installed built-in furniture. New apartment buyers inspect their newly built houses just before moving in to find deficiency. After inspection of fungal contamination or colonization in the built-in furniture and wall papers, there have been frequent claim of deficiency from fungal contamination. However, despite the frequent claims, there has been no domestic scientific data on fungal contaminants in built-in furniture of apartments. Therefore, this study was conducted to obtain case data of fungi causing problems in the built-in furniture of apartments in which fungal defects are claimed to exist.
2. Materials and methods
2.1. Fungal sampling and observation
Fungal samples were taken in 2016 from one of houses in a newly built apartment complex located in Goseong-gun, Gyeongsangnam-do. The house sampled had been claimed with fungal contamination before moving in by the owner. Thus, it was an empty new house with built-in furniture. To do sampling, we carefully inspected the house’s built-in furniture with naked eyes and a lope. We recognized the presence of fungi on built-in-furniture and interior made with wood board, plywood and MDF from kitchen, room 1, and the clothes wardrobe (). Consequently, a total of six spots were subject to samplings. Two samplings were from dress room, three samplings from kitchen, and one sampling from bedroom 1. Detail in sampling position is given in the legend of . Fungal samples were taken using sterile cotton swabs. Each swab was individually stored in 15 ml sterile plastic tube and moved to the laboratory for identification. Potato dextrose agar (PDA; BD, Detroit, MI, USA) media were smeared with each of the sampled cotton swabs and the smeared PDA plates were incubated for 3–5 days at 25 °C. Single spore isolation was performed on new PDA plates from the fungi grown out from the smeared PDA. Purely isolated fungal colonies were used for morphological and molecular analysis. To check the difference of colony pattern on the nutritional components of media, Czapek-dox yeast agar (CYA; Sigma-Aldrich, St. Luis, MO, USA), PDA, malt extract agar (MEA; BD), and oat meal agar (BD) were inoculated with the purely isolated fungi and incubated at 25 °C for 14 days. Colony morphology and colors were observed by naked eyes and photo was taken. The spores and mycelium of fungal colonies grown on PDA for 5 days were observed by a dissecting microscope (Model SZ61; Olympus, Tokyo, Japan) and an optical microscope (Model Axioskop40; Carl Zeiss, Oberkochen, Germany). The images of optical microscopes were documented.
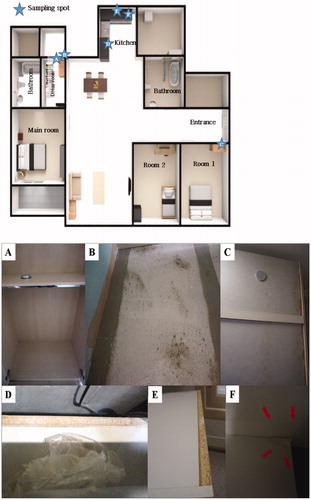
Figure 1. Sampling spot (top) in the floor plan of the sampled house and photos of the fungus contaminated built-in furniture (bottom). (A) The inside of closet in dress room (sampling spot1); (B) The back side of built-in furniture in dress room (sampling spot2); (C) The back side of closet in room 1 (sampling spot3); (D) Built-in furniture below the gas range in kitchen (sampling spot4); (E) The shelf of built-in furniture in kitchen (sampling spot5); (F) The built-in furniture in underneath of the sink in kitchen (sampling spot6).
2.2. DNA extraction and PCR amplification
Genomic DNA was extracted from PDA grown fungal mycelia cultured at 25 °C for 10 days according to the drilling method of Kim et al. [Citation10]. Calmodulin gene region and translation elongation factor 1-α (TEF-1α) gene region were analyzed by PCR using CL1 (5′-GARTWCAAGGAGGCCTTCTC-3′) and CL2A (5′-TTTTTGCATCATGAGTTGGA-3′) primers and TEF728 (5′-CATCGAGAAGTTCGAGAAGG-3′) and TEF1 (5′-GCCATCCTTGGAGATACCAGC-3′) primers according to the method of Tang et al. [Citation11]. The PCR amplified DNA was purified using the NAVIGenTM PCR Purification Kit (Navibiotech Corp., Cheonan, Korea) and its nucleotide sequence was determined in Macrogen Corp. (Seoul, Korea). The determined nucleotide sequences were searched through the BLAST program, a web-based program of the United State National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) for homologous fungal sequences.
2.3. Phylogenetic analysis
Nucleotide sequences of the calmodulin gene region and TEF-1α gene region of the fungi showing homology by the BLAST search program were designated as reference sequences. These reference sequences were downloaded from the DNA database of GenBank in NCBI. The determined nucleotide sequences of the isolated fungi in this study and reference sequences were subjected to multiple nucleotide sequence alignment using Cluster X program [Citation12]. From the multiple alignment data, a phylogenetic tree was drawn using MEGA 5.0 program [Citation13] to analyze the relationship between the isolated fungi and reference fungal species. Tree construction was performed using the neighbor joining method [Citation14] based on the nucleotide sequence of calmodulin gene region and TEF-1α gene region. 1000 bootstrapping analysis was carried out for clade reliability of phylogenetic tree.
2.4. Extracellular enzyme activity test
Extracellular enzyme activity of the isolated fungi was assayed on chromogenic media containing each substrate for β-glucosidase, avicelase, CM-cellulase, amylase, pectinase, xylanase, and proteinase [Citation15]. For the activity assay, the test fugal isolates were cultured on PDA for 14 days and their spores were collected and suspended in sterile water. The concentration of spore suspension of each fungal isolate was adjusted to 2 × 108 CFU/ml with sterile water and 5 μl of each spore suspension was inoculated in the center of the chromogenic medium. The medium inoculated with the test strain was cultured at 25 °C for 14 days, and then the presence of activities was verified by measuring the size of the clear zone formed on the chromogenic medium.
2.5. Inoculation test on wood chips
Four kinds of wood panels (Pine, Lauan, Jelutong, and medium density fiberboard (MDF)) were purchased from the market for building interior materials. The purchased panels were cut into wood chips of about 2.5 × 2.5 × 0.5 cm size and used for fungal inoculation experiments. The cut wood chip was immersed in 70% ethanol for 5 min for surface sterilization, air dried, and then irradiated with UV (254 nm) for 5 min for sterilization. After that, the wood chip was checked by culturing the UV sterilized wood chip on PDA to confirm that the wood chip is free of microorganisms. Of the six representative fungal isolates, four isolates with a response in the extracellular enzyme activity test were selected to investigate their ability of wood chip colonization. The selected fungal isolates were grown on PDA to produce spores, and then spore inocula were prepared by making a spore suspension at a concentration of 2 × 108 CFU/ml with sterile water. 10 μl of the inoculum of each isolate was spotted at the center of the surface of the sterilized wood chip. As a control, 10 μl of sterile water was spotted. Inoculation test was performed with three replicates. The inoculated wood chips were incubated in a thermo-hygrostat incubator at 25 °C and 30% humidity or 95% humidity condition for 10 days, respectively. Each of the fungal isolate colonized on wood chip was separated and confirmed to be the inoculated isolate.
3. Results and discussion
3.1. Isolation and identification
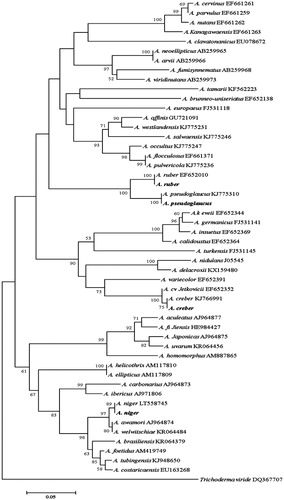
When we examined the fungal contamination on the built-in furniture of the apartment house, it seemed like that there was homogenous growing of a fungal species at each sampled spot. Thus, we could easily isolated and pure cultured fungi from the six sampling spots. All the isolates from each sampling spot showed the same colony morphology. One of the isolates was selected per each sampling spot and used as the representative culture for identification. Colony morphology of six representative isolates grown on CYA, PDA, MEA, and oat meal agar is given in . Blackish, dark brownish, yellowish, greenish, and whitish colors were shown in the colonies. These colors were actually observed when they were found on built-in furniture in . Interestingly, there was little difference in the mycelial texture, spreading pattern, and coloring when they were grown on the four different nutrient media. In addition, there was no identical colony pattern among the six isolates indicating they are likely different species. Isolate of the sampling spot 4 grew irregularly and easily differentiated form other five isolates. Aspergillus properties such as conidiophore of the erect hyphal branch which bulbously enlarges at an apex (columella) and gives rise to conidia were observed from four isolates. But their colony morphology was different. Thus, they were considered different species and coded as Aspergillus sp.1 DUCC6001, Aspergillus sp.2 DUCC6003, Aspergillus sp.3 DUCC6002, Aspergillus sp.4 DUCC6004. One isolate showed typical brush-like spore-bearing structures having flask-shaped phialides (called penicillin) from conidiophores which are simple or branched. Thus, it was coded as Penicillium sp.1 DUCC6000. The spores of spherical to lemon-shaped formed in long branching chains was observed and coded as coded as Cladosporium sp.1 DUCC6005. Colony morphology and microscopic observation of conidia and conidiophores of these six coded species were given and , respectively. One of the four Aspergillus species was typical shape of black A. niger [Citation16]. Since species in these fungal genera is hard to identify based only on morphology, molecular analysis was performed. Nucleotide sequence analysis of calmodulin gene through GenBank database search reveals that the four isolates of Aspergillus and one isolate of Penicillium from this study have 100% sequence identity with Aspergillus niger (LT558745), A. pseudoglacus (KJ775310), A. ruber(EF652010), and A. creber (KJ766991), and Penicillium commune (EF198594), respectively. In addition, nucleotide sequencing analysis of tef-1α gene from the six isolates in shows that one isolate of Cladosporium from this study has 100% sequence identity with that of C. perangustum (HM148379). The phylogenetic position of these six species is shown in , respectively. The six isolates from six sampling spots phylogenetically agreed with Aspergillus niger, A. pseudoglacus, A. ruber, and A. creber, C. perangustum and P. commune, respectively (). Thus, we identified Cladosporium sp.1 DUCC6000 as C. perangustum, Penicillium sp.1 DUCC6005 as P. commune, Aspergillus sp.1 DUCC6001 as A. niger, Aspergillus sp.2 DUCC6003 as A. pseudoglaucus, Aspergillus sp.3 DUCC6002 as A. ruber, and Aspergillus sp.4 DUCC6004 as A. creber. The determined nucleotide sequence of C. perangustum DUCC6005, P. commune DUCC6000, A. niger DUCC6001, A. ruber DUCC6002, A. pseudoglaucus DUCC6003, and A. pseudoglaucus DUCC6003 were registered on GenBank with accession numbers, MN619775, MN619776, MN619777, MN619788, MN619779, and MN619780, respectively.
Figure 2. Colony morphologies of the fungal isolates from six sampling spots formed on four different nutrient agar media after growing for 10 days at 25 °C.

Figure 3. Dissecting microscopic images of the six representative isolates from this study (scale bars = 1 mm). (A) Isolate of sampling spot3, Penicillium sp1 DUCC6000; (B) Isolate of sampling spot1, Cladosporium sp.1 DUCC6005; (C) Isolate of sampling spot2, Aspergillus sp.1 DUCC6001; (D) Isolate of sampling spot4, Aspergillus sp.2. DUCC6003; (E) Isolate of sampling spot5, Aspergillus sp.3 DUCC6002; (F) isolate of sampling spot6, Aspergillus sp.4 DUCC6004.
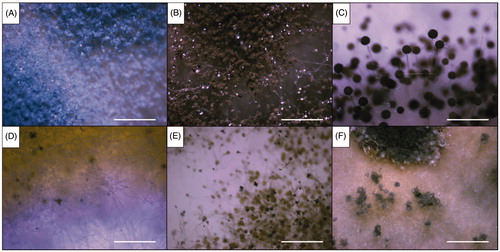
Figure 4. Optical microscopic images of the six representative isolates from this study (scale bars = 10 µm). (A) Isolate of sampling spot3, Penicillium sp1 DUCC6000; (B) Isolate of sampling spot1, Cladosporium sp.1 DUCC6005; (C) Isolate of sampling spot2, Aspergillus sp.1 DUCC6001; (D) Isolate of sampling spot4, Aspergillus sp.2. DUCC6003; (E) Isolate of sampling spot5, Aspergillus sp.3 DUCC6002; (F) isolate of sampling spot6, Aspergillus sp.4 DUCC6004.
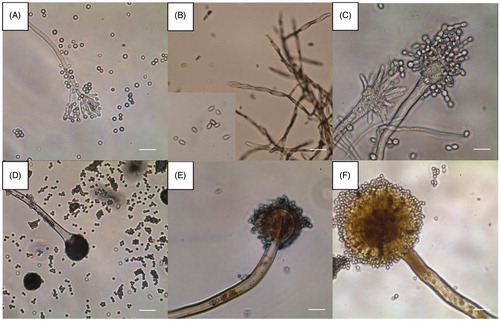
Figure 5. Phylogenetic tree for P. commune and related species based on neighbor-joining analysis of partial calmodulin gene region sequence using MEGA 5.0. The numbers at the nodes indicate the bootstrap support calculated for 1,000 repetitions. The scale bar indicates 0.05 substitutions per nucleotide position, outgroup is Trichoderma viride.
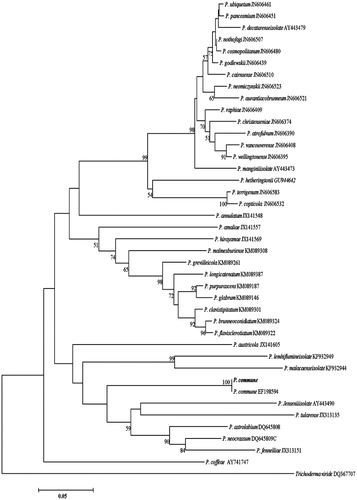
Figure 6. Phylogenetic tree for C. perangustum and related species based on neighbor-joining analysis of translation elongation factor 1 α (TEF - 1α) gene region sequence using MEGA 5.0. The numbers at the nodes indicate the bootstrap support calculated for 1,000 repetitions. The scale bar indicates 0.1 substitutions per nucleotide position, outgroup is Cercospora sojina. C. perangustum: Cladosporium perangustum DUCC6005.
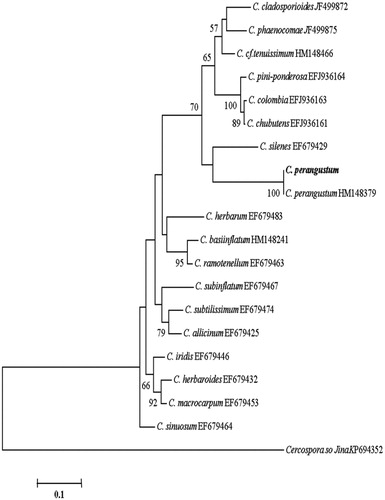
Figure 7. Phylogenetic tree for the four Aspergillus isolates and related species based on neighbor-joining analysis of partial calmodulin gene region sequence using MEGA 5.0. The numbers at the nodes indicate the bootstrap support calculated for 1,000 repetitions. The scale bar indicates 0.05 substitutions per nucleotide position, outgroup is Trichoderma viride. A. niger: Aspergillus niger DUCC6001, A. creber: Aspergillus creber DUCC6004. A. ruber: Aspergillus ruber DUCC6002, A. pseudoglucus: Aspergillus pseudoglaucus DUCC6003.
P. commune, A. ruber, A. pseudoglacus, and A. niger are known as food contaminants [Citation17–19]. A. niger is one of the most common species of the genus Aspergillus. It causes a disease called black mold on certain fruits and vegetables and is a common contaminant of food. It is ubiquitous in soil and commonly reported from indoor environments [Citation16]. Information on enzyme production is not available from A. pseudoglacus, A. creber, and C. perangustum. They were reported only by taxonomical studies. Recently, A. creber has been reported as unrecorded fungus in Korea [Citation20]. This species and A. niger are known to be function as opportunistic fungi [Citation21,Citation22]. C. perangustum has been reported to cause wood discoloration, corrosion, and disease in plants [Citation17,Citation23]. But colonization in built-in furniture environment is not much known in these six species.
3.2. Enzyme activity test results
Most of the elemental component of built-in furniture is wood components. Thus, the six identified fungal species’ potential to inhabiting on wood substrates was assessed by detecting activity of extracellular enzymes which are involved in degradation of wood components. For this assessment, seven kinds of enzymes were examined on chromogenic media (). A. creber showed protease and β-glucosidase activities. A. niger revealed β-glucosidase, while P. commune displayed xylanase and glucosidase activities. C. perangustum showed five kinds of enzyme activities including protease, amylase, avicelase (a form of cellulase), CM-celluase, and xylanase. A. pseudoglacus and A. ruber did not show any activities with the tested enzymes. Although protease was not detected on chromogenic media in this study, the analysis of A. niger genome reveals that this species contains serine proteases, aspartic proteases, metalloprotease, amino proteases, putative proteases, miscellaneous [Citation24]. In addition, the production of cellulase enzyme by a selected strain of A. niger isolated from deteriorated wood was reported [Citation25]. P. commune is known to produce pectinase [Citation26]. The report that A. ruber produced tannase under solid-state fermentation using the Jamun tree (Syzygium cumini) leaves [Citation27], suggests that it can colonize woody materials. Based on the results of and literature review, four out of the six species which showed at least one of the seven enzyme activities were selected and further analyzed with real wood materials.
Table 1. Extracellular enzyme activity test of fungi isolated from the built-in furniture.
3.3. Results of inoculation test on wood chips
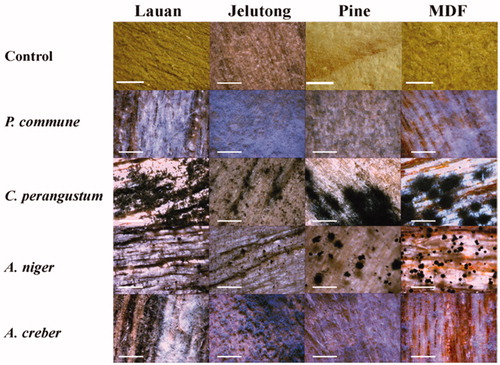
No fungus was re-isolated from the wood chips after sterilization. Thus, the sterilized wood chips were used for inoculation test. Control chips inoculated with sterile water showed no fungal growth ( and ). While, inoculation of P. commune, C. perangustum, A. niger, and A. creber showed that all of these species could grow on the surface of all the four kinds of lauan, madica, and pine wood chips and MDF plywood chips. The mycelial growth and spore formation of the four species were faster on MDF plywood chips than on the other types of wood chips. This observation is likely due to the increase of nutrients according to the organic substances which were added during the manufacture of MDF plywood. On the other hand, because of the characteristics produced by combining the structurally broken wood during processing, there is a physical gap. The gap could allow the inoculated fungi more easily penetrate into MDF plywood chips, and, in the end, may have helped they settle and grow. Therefore, MDF plywood has been shown to be more favorable than other wood for fungal growth [Citation28].
Figure 8. Results of inoculation test on wood chips at 25 °C and 30% humidity condition for 10 days (×45, scale bar = 1 mm). P. commune: Penicillium commune DUCC6000, C. perangustum: Cladosporium perangustum DUCC6005, A. niger: Aspergillus niger DUCC6001, and A. creber: Aspergillus creber DUCC6004.
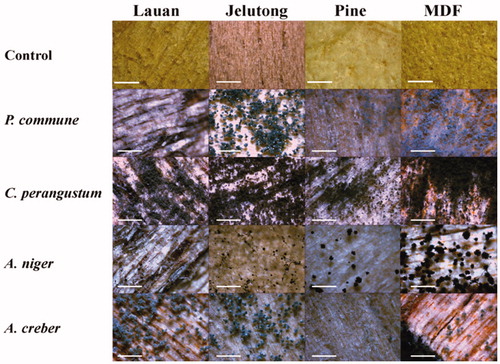
More spores and mycelia formed on the wood chips when the test fungal species were incubated at 95% humidity condition (). After incubation for 10 days, spore formation was more clearly induced at 95% humidity than at 30% humidity. A. creber and P. commune did not produce spores at 30% humidity. But the four species were still able to colonize the wood chips at 30% humidity condition, indicating they are good growers on the surface of wood even if at low humidity condition. Among the four fungal species, C. perangustum showed the most abundant and dense mycelial growth. In relation to the discoloration of wood, this species also broadly and darkly spreads its color with mycelia. Basically all the four species generated discoloration. The cause of discoloration is attributed to not by the secreted pigment but by the pigment deposited in the mycelia and spore cells. We could isolate again the four species from the inoculated chips. Based on the results of , we concluded that P. commune, C. perangustum, A. niger, and A. creber are able to colonize the built-in furniture in apartments.
Figure 9. Results of inoculation test on wood chips at 25 °C and 95% humidity condition for 10 days (×45, scale bar = 1 mm). P. commune: Penicillium commune DUCC6000, C. perangustum: Cladosporium perangustum DUCC6005, A. niger: Aspergillus niger DUCC6001, and A. creber: Aspergillus creber DUCC6004.
There have been studies on wood discoloration by staining fungi [Citation3] and decomposition of wood by wood decay [Citation25]. And building molds have been researched for building damage [Citation5,Citation6]. However, information on fungi related to built-in furniture is rarely available globally. Amid built-in furniture is getting popular in new apartment constructions in Korea, consumers’ demanding for eco-friendly furniture has been increasing. Consequently, the use of less toxic chemicals as the preservatives in the construction processing of furniture including built-in furniture has been required. This trend is likely resulting in the chance of fungal contamination in indoor environment. In this study we found out about the fungal species blooming in the built-in indoor furniture as well as the characteristics that can damage the furniture. Our work suggests it is necessary to manage fungal contamination in built-in furniture to reduce human impact and damage to furniture. The results of the present study are expected to serve as first case study on built-in furniture in apartment buildings. To resolve the fungal problems, further investigation should be needed to chase the origin of the fungal contamination in the built-in furniture in apartments by analyzing fungal contaminants before and after its installation.
Acknowledgements
The present research was conducted by the research fund of Dankook University in 2016.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Choi YJ. A study on improvement of indoor air quality in homes. The Seoul Institute; 2013.
- Reuss G, Disteldorf W, Gamer AO, et al. Formaldehyde. In: Ullmann’s Encyclopedia of Industrial Chemistry. Wiley‐VCH Verlag GmbH & Co. KgaA. 2002.
- Schwarzenegger A. Indoor air pollution in California. California EPA; 2005.
- Occupational Safety and Health administration. Formaldehyde. Standard number 1910.1048. U.S. Department of Labor; [Cited 2019 Sep 2].
- California Office of Environmental Health Hazard Assessment. Notice of adoption of air toxics hot spots program technical support document for the derivation of noncancer reference exposure levels and 6 RELS. 2008.
- Rumchev KB, Spicket JT, Bulsara MK, et al. Domestic exposure to formaldehyde significantly increases the risk of asthma in young children. Eur Respir J. 2002;20(2):403–406.
- Kim JY, Yoon JH, Lee CE, et al. Environmental hygienics. 9th ed. Seoul: KMS; 2017. p335–360.
- Moon HJ, Yoon YR, Park JW. An experimental study on mold germination and growth on wallpapers at different environmental conditions. J Archit Inst Korea. 2009;25(6):237–244.
- Kwon MH, Jang SK, Ryu JM, et al. A study on management of major indoor air pollutants by house type in Korea (I). NIER; 2009.
- Kim SH, Uzunovic A, Breuil C. Rapid detection of Ophiostoma piceae and O. quercus in stained wood by PCR. Appl Environ Microbiol. 1999;65(1):287–290.
- Tang LQ, Hyun MW, Yun YH, et al. New record of Mariannaea elegans var. elegans in Korea. Mycobiology. 2012;40(1):14–19.
- Thompson JD, Gibson TJ, Plewniak F, et al. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882.
- Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739.
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425.
- Yoon JH, Hong SB, Ko SJ, et al. Detection of extracellular enzyme activity in Penicillium using chromogenic media. Mycobiology. 2007;35(3):166–169.
- Rafai M, El-Yazid HA, Hassan A. Monograph on Aspergillus and aspergillosis in man, animals and birds. A guide for classification and identification of aspergilli, diseases caused by them, diagnosis and treatment. Academia; 2014.
- Kwon HW, Kim JY, Min SH, et al. Biochemical characterization of Agaricus bisporus dikaryon strains. Kor J Mycol. 2014;42(1):86–90.
- Sosa MJ, Córdoba JJ, Díaz C, et al. Production of cyclopiazonic acid by Penicillium commune isolated from dry-cured ham on a meat extract-based substrate. J Food Prot. 2002;65(6):988–992.
- Ge Y, Wang Y, Liu YX, et al. Two species of Aspergillus forming yellow cleistothecia popularly known as “golden flower” in dark brick tea of China. Mycosystema. 2015;34(2):186–195.
- Ahn GR, Choi MA, Kim JE, et al. A report of eighteen unrecorded fungal species in Korea. Kor J Mycol. 2017;45(4):292–303.
- Yun YH, Hyun MW, Suh DY, et al. Identification and characterization of Eurotium rubrum isolated from Meju in Korea. Mycobiology. 2009;37(4):251–257.
- Negri CE, Goncalves SS, Xafranski H, et al. Cryptic and rare Aspergillus species in Brazil: prevalence in clinical samples and in vitro susceptibility to triazoles. J Clin Microbiol. 2014;52(10):3633–3640.
- Choi MA, Park SJ, Ahn GR, et al. Identification and characterization of Paraconiothyrium brasiliense from garden plant Pachysandra terminalis. Kor J Mycol. 2014;2(4):262–268.
- Andersen MR, Salazar MP, Schaap PJ, et al. Comparative genomics of citric-acid-producing Aspergillus niger ATCC 1015 versus enzyme-producing CBS 513.88. Genome Res. 2011;21(6):885–897.
- Kassim EA. Cellulase enzyme from Aspergillus niger. Microbiol Immunol. 1982;26(6):449–454.
- Pitt JI, Cruickshank RH, Leistner L. Penicillium commune, P. camembertii, the origin of white cheese moulds, and the production of cyclopiazonic acid. Food Microbiology. 1986;3(4):363–371.
- Kumar R, Sharma J, Singh R. Production of tannase from Aspergillus ruber under solid-state fermentation using jamun (Syzygium cumini) leaves. Microbiol Res. 2007;162(4):384–390.
- Jurjevic Z, Peterson SW, Horn BW. Aspergillus section Versicolores: nine new species and multilocus DNA sequence based phylogeny. IMA Fungus. 2012;3(1):59–79.
