Abstract
Talaromyces halophytorum sp. nov. was isolated from the roots of halophyte Limonium tetragonum collected from Seocheon-gun, Korea in November 2015. It showed a slow growth on yeast extract sucrose agar at 25 °C, no growth at 4 °C or 37 °C and produced smooth-walled and globose to sub-globose conidia. T. halophytorum is phylogenetically distinct from the other reported Talaromyces species of section Trachyspermi based on multi-locus sequence typing results using partial fragments of β-tubulin, calmodulin, ITS, and RNA polymerase II genes.
1. Introduction
The genus Talaromyces was established by Benjamin (1955) as a name for a number of Penicillium species that could produce a sexual state. The genus was characterized by producing soft ascocarps with cleistothecial walls that have multiple layers of interwoven hyphae and typically yellow ascomata with ovate to globose asci with spiny ascospores [Citation1–3]. Based on phenotypic, extrolite, phylogenetic data and the concept of one fungus one name, Samson et al. [Citation4] transferred the majority of accepted species of Penicillium subg. Biverticillium to Talaromyces. In 2014, a total of 88 species were accepted in the monograph of the genus. These species were classified in seven well-defined sections, namely Talaromyces, Bacillispori, Helici, Purpurei, Trachyspermi, Subinflati, and Islandici [Citation3]. Subsequent to the monograph of Yilmaz et al. [Citation3], 73 new Talaromyces species such as T. amyrossmania, T. heiheensis, T. minnesotensis, T. rubrifaciens, and many more have been described from all over the world [Citation5–10].
Talaromyces species are thoroughly associated with human life since they could be used as potential anti-cancer [Citation11], anti-fungal [Citation12], and anti-Trypanosoma [Citation13] agents. Talaromyces species have also received considerable attention as biotechnological resources such as purpactins (produce by T. purpurogenus as inhibitors of acyl–coenzyme A), anti-influenza virus polyketides [Citation14,Citation15], antibiotics [Citation12], food dyes [Citation16], and exoenzymes such as dextranases, cellulases, glucoamylases, and chitinases [Citation17–20]. Other Talaromyces species, like T. marneffei, are isolated from clinical specimens [Citation21] and can cause fatal mycoses in immunocompromised individuals [Citation22].
Limonium tetragonum (Thunb.) A. A. Bullock (Plumbaginaceae) is a species of salt-tolerant plants that grows in saline environments such as coastal sand dunes, salt marsh, and muddy seashores throughout the western coastal area of Korea [Citation23]. Roots and leaves of this plant are widely used as edible vegetables in Korea. It has been reported that it has antioxidant and anti-cancer properties due to the presence of numerous bioactive materials [Citation24,Citation25].
In the present study, we report a new Talaromyces species in section Trachyspermi isolated from L. tetragonum.
2. Materials and methods
2.1. Collection, strain isolation, and preservation
L. tetragonum plants were collected from the coast of Seocheon-gun, Chungcheongnam-do, Korea in November 2015. Samples were transported to the laboratory and processed for isolation of endophytic fungi within 24 h. First, the plant material was rinsed with tap water to remove dust and debris. The stem and root were then cut into small pieces with a sterilized blade under aseptic conditions. Five tissue segments were randomly selected from each part. These plant tissues were surface-sterilized by consecutive immersions in 70% ethanol for 30 s and in 1% sodium hypochlorite (NaOCl; Duksan, Ansan, Korea) for 1 min. These samples were then rinsed with sterile water three times and allowed to surface dry on filter paper. The sets of five tissue segments from stem and root were placed on potato dextrose agar (PDA; Difco, Sparks, MD, USA) plate and incubated at 25 °C. As the fungus grew from the tissue segment, newly emerging hyphal tips were transferred onto new PDA plates. These plates were incubated at 25 °C to obtain single hyphae isolate. Pure cultures of fungal strains were preserved on PDA slant at 4 °C. A representative strain (WLT07) was deposited in the Korean Agricultural Culture Collection, National Institute of Agricultural Science, Rural Development Administration, Wanju, South Korea (KACC 48127) to be used for further studies.
2.2. Morphological analysis
Strain WLT07 was morphologically studied on different media under different growth conditions. The strain was inoculated onto malt extract agar (MEA; Oxoid, Hampshire, UK), Czapek yeast extract agar (CYA; Difco), yeast extract sucrose agar (YES), oatmeal agar (OA; Difco), and creatine sucrose agar (CREA) at three points on 90-mm Petri dishes and incubated at 25 °C in the dark for 7 days. All media were prepared as described by Visagie et al. [Citation26]. Additional CYA plates were incubated at 4 °C and 37 °C for 7 days in the dark. After incubation, diameters of colonies on each medium were measured. The density of sporulation, obverse and reverse colony colors, and the production of soluble pigments were noted. Fungal colonies were photographed with a Canon EOS 400 D camera (Tokyo, Japan). Morphological characterization was performed by observing the slides prepared from MEA using light microscopy (DE/Axio Imager.A1, Carl Zeiss, Göttingen, Germany). Lactic acid was used as a mounting fluid and a drop of ethanol was added to remove excess conidia. Specimen images were acquired using AxioVision LE64 software (Carl Zeiss, Oberkochen, Germany). Figure plates were prepared with Photoshop CS2 (Adobe, San Jose, CA, USA).
2.3. Multi-gene sequencing and phylogenetic analysis
To extract genomic DNA, strain WLT07 was grown in malt extract broth medium (MEB; Oxoid, Hampshire, UK) on an orbital shaker for 2–4 days at 25 °C. Its fungal mycelia were harvested by filtration and transferred to sterile 1.5 mL tubes. These samples were frozen at −70 °C, lyophilized, and finely ground. DNA was extracted using a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The internal transcribed spacer (ITS) region of the ribosomal DNA was amplified using ITS1 and ITS4 primers [Citation27]. In addition, three nuclear protein genes were amplified (partial β-tubulin (benA) gene was amplified using Bt2a and Bt2b [Citation28]; partial DNA-dependent RNA polymerase II second largest subunit (Rpb2) gene was amplified using RPB2-5F and RPB2-7CR [Citation29]; and partial calmodulin (CaM) gene was amplified using CMD5 and CMD6 [Citation30]). PCRs were performed in 25 μL reaction tubes containing 2.0 μL DNA template, 0.5 μL of each forward and reverse primers (10 μmol L−1), 0.5 µL Taq DNA polymerase (Bioneer, Daejeon, Korea), 0.5 µL of each dNTP, 2.5 µL 10× PCR reaction buffer, and 18.5 μL of sterile double-distilled water. Thermal cycling conditions were as follows: initial denaturation at 94 °C for 3 min; 35 cycles of 94 °C for 40 s, 54 °C for ITS, 58 °C for benA, 55 °C for CaM, or 48 °C for RPB2 for 60 s, and 72 °C for 2 min; and a final elongation step of 72 °C for 10 min. PCR products were purified and sequenced by Macrogen (Seoul, Korea). Nucleotide sequences obtained were searched using BLASTn available in the GenBank database (http://www.ncbi.nlm.nih.gov/BLAST/) to obtain the most likely taxonomic designations for the strain (). Evolutionary matrices for the maximum likelihood, neighbor-joining, and maximum parsimony were constructed using Kimura’s two-parameter model [Citation31]. Phylogenetic tree topology was inferred by the maximum likelihood, neighbor-joining, and maximum-parsimony method using MEGA6 with bootstrap values based on 1000 replications [Citation32].
Table 1. Accession numbers of fungal strains used for the phylogenetic analysis.
3. Results
3.1. Morphological characterization
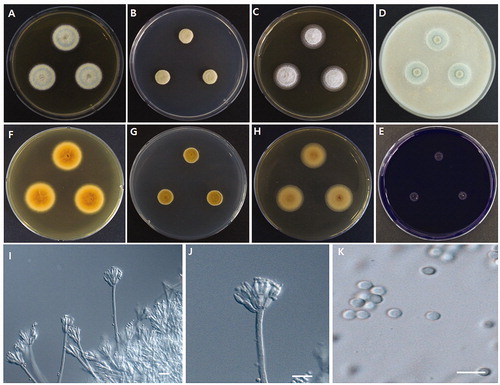
Photomicrographs of morphological structures of the WLT07 strain are shown in . Detailed morphological characters are described in the Taxonomy section (Section 3.3).
3.2. Phylogenetic analysis
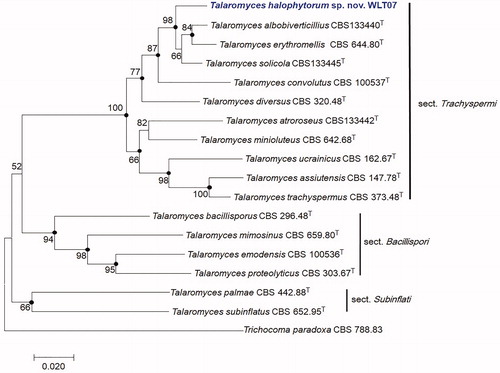
Sizes of PCR amplicons were 551 bp for ITS, 423 bp for BenA, 479 bp for CaM, and 1088 bp for RPB2. Sequences obtained from strain WLT07 were deposited in the NCBI database (GenBank accession numbers: MH725786 for ITS, MH729367 for BenA, MK111426 for CaM, and MK111427 for RPB2). Phylogenetic analysis based on the maximum-likelihood methods of combined ITS, BenA, CaM, and RPB2 sequences showed that strain WLT07 is closely related with type strains of T. solicola CBS 133445 (96.3% homology), T. albobiverticillius CBS 133440 (93.5%), and T. erythromellis CBS 644.80 (93.4%). Neighbor-joining and maximum-parsimony phylograms were also constructed to determine the exact taxonomic position of the strain and the bootstrap values are indicated at the nodes in the maximum-likelihood phylogenetic tree. Filled circles indicated that corresponding nodes were also recovered in trees generated with the neighbor-joining and maximum parsimony algorithms (). The phylogenetic tree revealed that the phylogenetic position of WLT07 was clearly separated from T. solicola, T. albobiverticillius, and T. erythromellis. Thus, WLT07 was phylogenetically distinct from other species of Talaromyces.
Figure 2. Maximum-likelihood phylogenetic tree based on combined ITS, BenA, CaM, and RPB2 genes of Talaromyces section Trachyspermi species including Talaromyces halophytorum sp. nov., WLT07. Filled circles indicate that corresponding nodes are also recovered in the trees generated with the maximum-parsimony and neighbor joining algorithms. Trichocoma paradoxa was included as an outgroup. Bootstrap analysis was performed with 1000 replications. T indicates the type strain of the species. Bar, 0.02 substitutions per nucleotide position.
3.3. Taxonomy
Talaromyces halophytorum, Y. H. You and S. B. Hong, sp. nov. ().
Mycobank: MB830295.
Etymology: ha.lo.phy.to’rum, N.L. gen. pl. n. halophytorum of halophytes.
In: Talaromyces section Trachyspermi.
Typus: KACC 48127 (NIBRFGC 000501933).
Gene sequence: ITS= MH725786; BenA=MH729367; CaM=MK111426; RPB2 = MK111427.
Colony diam., 7 d (mm): MEA 24–25; CYA 12–13; CYA 4 °C and 37 °C No growth; YES 18–20; OA 15–6; CREA 3–5.
Colony characteristics: MEA, 25 °C, 7 days: colony greenish gray in color, reverse brownish orange, raised in center, concentric, margins narrow (1–2 mm), low, entire plane; texture floccose; sporulation dense, conidia en masse grayish green; exudates clear, sometimes orange; soluble pigment absent (). CYA, 25 °C, 7 days: colony gray in color, reverse brownish orange to yellowish brown, raised at margins, occasionally cleaved (or cracked) in the middle, texture mainly velvety, occasionally floccose; sporulation dense, conidia en mass grayish green; soluble pigment, and exudate absent (). YES, 25 °C, 7 days: colony white in color, backside light orange, flat, slightly concentrically sulcate, margins entire; texture floccose; sporulation sparse; soluble pigment and exudate absent (). OA, 25 °C, 7 days: colony greenish to green, raised at center, texture velvety, especially near center; sporulation moderately to dense; soluble pigment and exudates absent (). CREA, 25 °C, 7 days: very weak growth, acid production absent ().
Micromorphology: Conidiophores biverticillate, 70–180 × 3–4 µm. Subterminal branches absent. Stipes smooth-walled. Phialides acerose, 3–5 per metula, and 6.5–11 × 2.5–3 µm in size. Metulae arranged in verticils of 5–8, and 6.5–11 × 2.5–4 µm in size (). Conidia globose to subglobose, 2.5–3.5 × 2–3 µm in diameter, smooth-walled (). Ascomata absent.
Distinguishing characters: Talaromyces halophytorum is characterized by its slow growth on CYA and no growth on CYA at 4 °C or 37 °C. Conidiophores are biverticillate and it produces smooth-walled globose to sub-globose conidia. Phylogenetically, it is closely related to T. silicola, T. erythromellis, and T. albobiverticillius. However, it is differentiated from these three species in morphology. T. erythromellis produces symmetrical subterminal branches and smooth-walled sub-globose to ellipsoidal conidia, whereas T. solicola and T. albobiverticillius produce rough-walled globose to sub-globose shaped conidia.
4. Discussion
A phylogenetic approach based on multiple genes [Citation33] (ITS, BenA, CaM, and RPB2) was applied to study the relationship of T. halophytorum in Talaromyces section Trachyspermi. Our results inferred from the phylogenetic analysis of combined ITS, BenA, CaM, and RPB2 sequences indicated that T. albobiverticillius, T. erythromellis, and T. solicola formed a group [Citation3,Citation16]. Phylogenetic trees revealed that strain WLT07 is distinct from other known Talaromyces species. This result is confirmed by results of neighbor-joining, maximum parsimony, and maximum-likelihood phylogenetic trees (). Yaguchi et al. [Citation34] introduced Talaromyces section Trachyspermi (as “trachyspermus”) based on ubiquinone systems that overrode the traditional morphology-based classification of Talaromyces. Section Trachyspermi was established for species that grew restrictedly on CYA, YES, and DG18, slightly faster on MEA, and poorly on CREA. Conidiophores are biverticillate and ascomata, if present, have a cream white or yellow color [Citation3]. T. halophytorum strain WLT07 in this group is supported by similarities in its morphological characters including restricted growth on CYA, YES, slightly faster growth on MEA, poor growth on CREA, biverticillated conidiophores, smooth-walled conidia, and globose to sub-globose shape (). Despite these similarities, T. halophytorum could be distinguished from T. solicola, T. erythromellis, and T. albobiverticillius by distinctive phenotypic characters such as colors on CYA, CYA soluble pigment, conidial ornamentation, and shape. The reverse side color on CYA of T. halophytorum was brownish orange to yellowish brown, whereas that of T. solicola was reddish brown to dark brown, that of T. erythromellis was dark red to brownish red, and that of T. albobiverticillius was grayish red-brown. Only T. albobiverticillius produced red pigments on CYA. Another notable feature of T. halophytorum was that it produced biverticillated conidiophores and smooth-walled globose to sub-globose conidia, whereas T. erythromellis produced symmetrical subterminal branches and smooth-walled sub-globose to ellipsoidal conidia and T. solicola and T. albobiverticillius produced rough conidia with globose to sub-globose shape [Citation3,Citation16,Citation35]. In conclusion, our data show that this strain represents a new species of Talaromyces in section Trachyspermi. Talaromyces halophytorum sp. nov. is proposed as its name.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Benjamin CR. Ascocarps of Aspergillus and Penicillium. Mycologia. 1955;47(5):669–687.
- Pitt JI. The genus Penicillium and its teleomorphic states Eupenicillium and Talaromyces. London, UK: Academic Press; 1979.
- Yilmaz N, Visagie CM, Houbraken J, et al. Polyphasic taxonomy of the genus Talaromyces. Stud Mycol. 2014;78:175–341.
- Samson RA, Yilmaz N, Houbraken J, et al. Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Stud Mycol. 2011;70:159–183.
- Jiang XZ, Yu ZD, Ruan YM, et al. Three new species of Talaromyces sect. Talaromyces discovered from soil in China. Sci Rep. 2018;8(1):4932.
- Crous PW, Wingfield MJ, Burgess TI, et al. Fungal Planet description sheets 625–715. Persoonia. 2017;39:460–461.
- Barbosa RN, Bezerra JD, Souza-Motta CM, et al. New Penicillium and Talaromyces species from honey, pollen and nests of stingless bees. Antonie Van Leeuwenhoek. 2018;13:1–30.
- Su L, Niu YC. Multilocus phylogenetic analysis of Talaromyces species isolated from curcurbit plants in China and description of two new species, T. curcurbitiradicus and T. endophyticus. Mycologia. 2018;110(2):375–386.
- Varriale S, Houbraken J, Granchi Z, et al. Talaromyces borbonicus sp. nov., a novel fungus from biodegraded Arundo donax with potential abilities in lignocellulose conversion. Mycologia. 2018;27:1–9.
- Rajeshkumar KC, Yilmaz N, Marathe SD, et al. Morphology and multigene phylogeny of Talaromyces amyrossmaniae, a new synnematous species belonging to the section Trachyspermi from India. Mycokeys. 2019;45:41–56.
- Li H, Huang H, Shao C, et al. Cytotoxic norsesquiterpene peroxides from the endophytic fungi Talaromyces flavus isolated from the mangrove plant Sonneratia apetala. J Nat Prod. 2011;75:1230–1235.
- Zhai MM, Li J, Jiang CX, et al. The bioactive secondary metabolites from Talaromyces species. Nat Prod Bioprospect. 2016;6(1):1–24.
- Cota BB, Rosa LH, Caligiorne RB, et al. Altenusin, a biphenyl isolated from the endophytic fungus Alternaria sp., inhibits trypanothione reductase from Trypanosoma cruzi. FEMS Microbiol Lett. 2008;285(2):177–182.
- Tomoda H, Nishida H, Masuma R, et al. Purpactins, new inhibitors of acyl-CoA: cholesterol acyltransferase produced by Penicillium purpurogenum. I. Production, isolation and physico-chemical and biological properties. J Antibiot. 1991;44(2):136–143.
- Wang H, Wang Y, Wang W, et al. Antiinfluenza virus polyketides from the acid-tolerant fungus Penicillium purpurogenum JS03-21. J Nat Prod. 2011;74(9):2014–2018.
- Frisvad JC, Yilmaz N, Thrane U, et al. Talaromyces atroroseus, a new species efficiently producing industrially relevant red pigments. PLoS One. 2013;8(12):e84102.
- Erhardt FA, Stammen S, Jördening HJ. Production, characterization and (co-) immobilization of dextranase from Penicillium aculeatum. Biotechnol Lett. 2008;30(6):1069–1073.
- Morozova VV, Gusakov AV, Andrianov RM, et al. Cellulases of Penicillium verruculosum. Biotechnol J. 2010;5(8):871–880.
- Volkov PV, Rozhkova AM, Semenova MV, et al. Comparative study of biochemical properties of glucoamylases from the filamentous fungi Penicillium and Aspergillus. Biochemistry Moscow. 2013;78(10):1180–1189.
- Binod P, Pusztahelyi T, Nagy V, et al. Production and purification of extracellular chitinases from Penicillium aculeatum NRRL 2129 under solid-state fermentation. Enzyme Microb Technol. 2005;36(7):880–887.
- Li L, Chen K, Dhungana N, et al. Characterization of clinical isolates of Talaromyces marneffei and related species, California, USA. Emerg Infect Dis. 2019;25(9):1765–1768.
- Hien TV, Loc PP, Hoa NTT, et al. First case of disseminated Penicilliosis marneffei infection among patients with acquired immunodeficiency syndrome in Vietnam. Clin Infect Dis. 2001;32:78–80.
- Ihm BS, Lee JS, Kim JW. Costal vegetation on the western, southern, and eastern coasts of South Korea. J Plant Biol. 2001;44(3):163–170.
- Kong CS, Um YR, Lee JI, et al. Inhibition effects of extracts and its solvent fractions isolated from Limonium tetragonum on growth of human cancer cells. Kor Soc Biotechnol Bioeng J. 2008;23:177–182.
- Lee JI, Kong CS, Jung ME, et al. Antioxidant activity of the halophyte Limonium tetragonum and its major active components. Biotechnol Bioproc E. 2011;16(5):992–999.
- Visagie CM, Houbraken J, Frisvad JC, et al. Identification and nomenclature of the genus Penicillium. Stud Mycol. 2014;78:343–371.
- White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. p. 315–322.
- Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous Ascomycetes. App Environ Microbiol. 1995;61(4):1323–1330.
- Liu YJ, Whelen S, Hall BD. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol Biol Evol. 1999;16(12):1799–1808.
- Hong SB, Cho HS, Shin HD, et al. Novel Neosartorya species isolated from soil in Korea. Int J Syst Evol Microbiol. 2006;56(2):477–486.
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111–120.
- Tamura K, Stecher G, Peterson D, et al. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729.
- Taylor JW, Jacobson DJ, Kroken S, et al. Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol. 2000;31(1):21–32.
- Yaguchi T, Someya A, Udagawa SI. A reappraisal of intrageneric classification of Talaromyces based on the ubiquinone systems. Mycoscience. 1996;37(1):55–60.
- Visagie CM, Jacobs K. Three new additions to the genus Talaromyces isolated from Atlantis sandveld fynbos soil. Persoonia. 2012;28(1):14–24.