Abstract
Alternaria alternata JS-1623 is an endophytic fungus isolated from a stem tissue of Korean fir, Abies koreana. Ethyl acetate extracts of culture filtrates exhibited anti-inflammatory activity in LPS induced microglia BV-2 cell without cytotoxicity. Here we report a 33.67 Mb sized genome assembly of JS-1623 comprised of 13 scaffolds with N50 of 4.96 Mb, and 92.41% of BUSCO completeness. GC contents were 50.97%. Of the 11,197 genes annotated, gene families related to the biosynthesis of secondary metabolites or transcription factors were identified.
The genus Alternaria are well known as plant-pathogenic and/or saprophytic fungi and are capable of producing various secondary metabolites including mycotoxins [Citation1–3]. These mycotoxins may play an important role in the fungal pathogenesis toward plant. Unlike plant pathogenic Alternaria species, endophytic Alternaria species have been also found to be prolific producer of secondary metabolites, but these are mostly beneficial having anti-microbial [Citation4,Citation5], anti-inflammation [Citation4] and anti-cancer [Citation6] as well as anti-viral activities [Citation7]. A. alternata is also known as both plant pathogen and endophytes and produce a number of potential bioactive metabolites [Citation5,Citation7,Citation8]. Although a number of studies have been focused on analysis and efficacy of the metabolites, little is known about the genes-related to production of substances in this fungus.
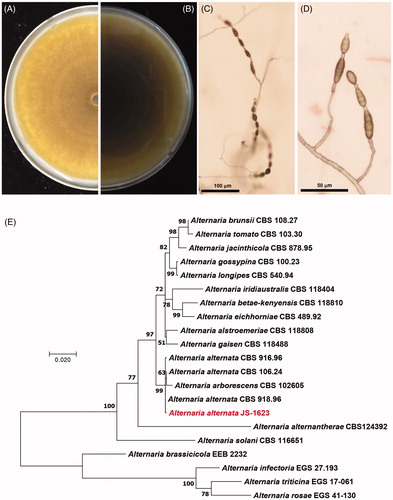
A. alternata strain JS-1623 was isolated from a stem tissue of Korean fir, Abies koreana collected from Mt. Gaya, Seongju (35° 49′ 21″N, 128° 7′ 25″E), South Korea in 2014, following the procedure described previously [Citation9]. This strain was deposited in Wildlife Genetic Resources Bank at the National Institute of Biological Resources (Incheon, Korea) with the accession number of NIBRGR0000180467. Ethyl acetate extract of this strain showed significant nitric oxide reduction activity in LPS-stimulated microglial BV-2 cell, while did not exhibit cytotoxicity () as tested according to the methods reported previously [Citation10]. Colonies were dark yellow on the front () and dark brown on the reverse side () of the PDA plate. On Potato Carrot Agar [Citation11], JS-1623 generated elongated conidial chains on top of straight or curved and short or long conidiophores which are dark brown in color. Conidia are septate, obclavate and long ellipsoide (). Using MEGA X program [Citation12], maximum likelihood analysis based on the concatenated GAPDH (MN633402), tef1 (MN633409) and alta1 (MN633400) sequences placed the JS-1623 isolate within a clade comprising A. alternata ().
Figure 1. Anti-inflammatory activity (A) and cytotoxicity (B) of Alternaria alternata JS-1623. (A) Effects of ethyl acetate extracts of JS-1623 culture filtrates on extracellular nitric oxide level of LPS induced microglial BV-2 cell. Microglial BV-2 cells were pretreated with extracts of JS-1623 at final concentration of 50 ppm for 3 h before stimulation with LPS (1 μg/ml) for 24 h. Griess reagent was used to detect the extracellular nitrite. (B) Effects of extracts on cell viability. BV-2 cells were treated with 50 ppm extracts for 24 h before viability was measured. Data obtained from three biological replicates are shown as the means ± s.d. values. Bars with the same letter are not significantly different (Duncan’s multiple comparisons test, *p < 0.05).
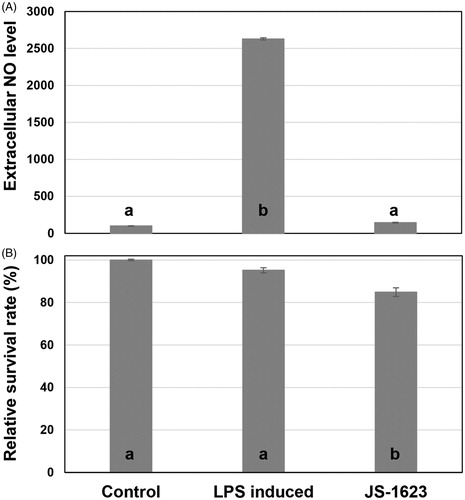
Figure 2. Morphological characteristics and phylogenetic analysis of Alternaria alternata JS-1623. (A) Ten-day-old colony on the upper side of PDA. (B) Light brown pigmentation on the reverse side of PDA. Microscopic picture of conidial chain on conidiophore at (C) × 100 and (D) × 200 magnification. (E) Maximum likelihood (ML) tree was constructed by MEGA X. The phylogenetic tree was generated from concatenated GAPDH (MN633402), tef1 (MN633409) and alta1 (MN633400) sequences with JS-1623. Bootstrap = 2000. The numbers at each node represent bootstrap values.
Genomic DNA was extracted from young mycelia grown in potato dextrose broth for two days at 25 °C by shaking at 120 rpm using Qiagen DNeasy plant mini kit (Valencia, CA), according to the manufacturer’s instructions. Library preparation and genome sequencing was performed at Theragen Etex Bio Institute, Suwon, Korea, following the strategies described previously [Citation13]. The single-molecule real-time (SMRT) sequencing library with a fragment size of 20 kb was prepared using SMRTbell Template Prep Kit (Pacific Biosciences, Menlo Park, CA). A short-insert paired-end (PE) library with fragment size of 350 bp, and a long-insert mate-pair (MP) library with fragment size of 10 kb were constructed using TruSeq Nano DNA sample prep kit and Nextera mate pair library prep kit (Illumina, San Diego, CA), respectively.
A total of 637,628 reads with an average size of 6.3 kb were obtained from PacBio Sequel platform. These reads were assembled into 47 contigs with a total length of 3,37,47,404 bp and an N50 value of 2.84 Mb using the overlap-layout-consensus (OLC) algorithm [Citation14–16]. Two short-read sequencing data sets, i.e., about 4.80 Gb with a Q30 of 83.42% a PE library, and 6,406,552 high-quality reads from a MP library after filtering with NextClip v1.3 [Citation17] were obtained using 101 cycles of paired-end sequencing on Illumina HiSeq 2000 platform. These short reads were assembled using SOAPdenovo v2.04 to 65 contigs with a total length of 33,342,242 bp and an N50 value of 2.02 Mb. The two assemblies were merged with HaploMerger2 [Citation18] to 21 contigs, followed by scaffolding and gap filling using SSPACE-Standard v3.0 [Citation19], SSPACE-LongRead v1.1 [Citation20], and GapFiller v1.10 with default parameter [Citation21], respectively. Finally, from the 13.74 Gb (392.57-fold coverage) of genome sequences, the reads containing the nuclear genome were assembled into 13 scaffolds with a total length of 33,672,707 bp and an N50 value of 4.96 Mb (). The genome assembly was validated using BUSCO v. 3.0.2b, which showed 268 (92.41%) BUSCOs as complete without any complete and duplicated ones out of 290 fungal gene set [Citation22]. The GC content of the assembled genome was 50.97%.
Table 1. Genome assembly and annotation statistics of Alternaria alternata JS-1623.
A total of 11,514 protein-coding gene models were predicted by AUGUSTUS v.3.2.1 [Citation23], 97.25% (11,197 genes) of which were functionally annotated through BLAST against UniProt or NCBI nonredundant (nr) and InterPro scan. Biosynthetic gene clusters (BGCs) with 11 polyketide synthase and 12 non-ribosomal protein synthetase were identified through antiSMASH v5.0 [Citation24]. Functional prediction was further conducted to find 307 transcription factor genes by the Fungal Transcription Factor Database (FTFD) v1.0 [Citation25], 147 cytochrome P450 genes by the Fungal Cytochrome P450 Database v1.0 [Citation26]. About 2.2% of genome were composed of repetitive sequences including retrotransposons, DNA transposons, simple sequence repeats, and unclassified repeats through the combining of RebBase [Citation27,Citation28] search and de novo identification data using RepeatModeler 1.x.x and RepeatMasker 4.0.x [Citation29]. A total of 349 non-coding RNAs, comprised of 46 rRNA from BLASTn search against NCBI, 147 tRNA through tRNAscan-SE [Citation30], 42 microRNA and 114 small nucleolar RNA through Infernal search against Rfam 11.0 [Citation31], were also identified.
This whole-genome sequence of A. alternata will provide a good basis to identify genes related with biosynthesis of valuable secondary metabolites as well as genes involved in virulence of pathogenic strains by genome comparison. The genome of A. alternata JS-1623 reported here has been deposited in GenBank under accession no. VZUP00000000. The version described in this article is version VZUP01000000 (https://www.ncbi.nlm.nih.gov/nuccore/ VZUP00000000.1).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Lee HB, Patriarca A, Magan N. Alternaria in food: ecophysiology, mycotoxin production and toxicology. Mycobiology. 2015;43(2):93–106.
- Luo H, Tao YQ, Fan XY, et al. Identification and characterization of Alternaria iridiaustralis causing leaf spot on iris ensata in China. Mycobiology. 2018;46(2):168–171.
- Ostry V. Alternaria mycotoxins: an overview of chemical characterization, producers, toxicity, analysis and occurrence in foodstuffs. World Mycotoxin J. 2008;1(2):175–188.
- Moharram AM, Zohri AA, Omar HM, et al. In vitro assessment of antimicrobial and anti-inflammatory potential of endophytic fungal metabolites extracts. Eur J Biol Res. 2017;7:234–244.
- Wang XZ, Luo XH, Xiao J, et al. Pyrone derivatives from the endophytic fungus Alternaria tenuissima SP-07 of Chinese herbal medicine Salvia przewalskii. Fitoterapia. 2014;99:184–190.
- Shen L, Tian SJ, Song HL, et al. Cytotoxic tricycloalternarene compounds from endophyte Alternaria sp. W-1 associated with Laminaria japonica. Mar Drugs. 2018;16:pii: E402.
- Bashyal BP, Wellensiek BP, Ramakrishnan R, et al. Altertoxins with potent anti-HIV activity from Alternaria tenuissima QUE1Se, a fungal endophyte of Quercus emoryi. Bioorg Med Chem. 2014;22(21):6112–6116.
- Li DM, Zhang YH, Ji HX, et al. Tricycloalternarene derivatives from endophytic fungus Alternaria tenuissima SY-P-07. Nat Prod Res. 2013;27(20):1877–1881.
- Nguyen HT, Kim S, Yu NH, et al. Antimicrobial activities of an oxygenated cyclohexanone derivative isolated from Amphirosellinia nigrospora JS-1675 against various plant pathogenic bacteria and fungi. J Appl Microbiol. 2019;126(3):894–904.
- Lee C, Kim S, Li W, et al. Bioactive secondary metabolites produced by an endophytic fungus Gaeumannomyces sp. JS0464 from a maritime haplophyte Phragmites communis. J Antibiot. 2017;70(6):737–742.
- Simmons EG. Alternaria: an identification manual. Urecht (Netherlands): Centraalbureau voor Schimmelcultures; 2007.
- Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549.
- Jeon J, Park SY, Kim JA, et al. Draft genome sequence of Amphirosellinia nigrospora JS-1675, an endophytic fungus from Pteris cretica. Microbiol Resour Announc. 2019;8(20):pii: e00069-19.
- Flicek P, Birney E. Sense from sequence reads: methods for alignment and assembly. Nat Methods. 2009;6(S11):S6–S12.
- Miller JR, Koren S, Sutton G. Assembly algorithms for next-generation sequencing data. Genomics. 2010;95(6):315–327.
- Schatz MC, Delcher AL, Salzberg SL. Assembly of large genomes using second-generation sequencing. Genome Res. 2010;20(9):1165–1173.
- Leggett RM, Clavijo BJ, Clissold L, et al. NextClip: an analysis and read preparation tool for Nextera Long Mate Pair libraries. Bioinformatics. 2014;30(4):566–568.
- Huang S, Chen Z, Huang G, et al. HaploMerger: reconstructing allelic relationships for polymorphic diploid genome assemblies. Genome Res. 2012;22(8):1581–1588.
- Boetzer M, Henkel CV, Jansen HJ, et al. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27(4):578–579.
- Boetzer M, Pirovano W. SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinformatics. 2014;15(1):211.
- Nadalin F, Vezzi F, Policriti A. GapFiller: a de novo assembly approach to fill the gap within paired reads. BMC Bioinformatics. 2012;13(S14):S8.
- Waterhouse RM, Seppey M, Simao FA, et al. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. 2018;35(3):543–548.
- Stanke M, Steinkamp R, Waack S, et al. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 2004;32 (Web Server):W309–W312.
- Blin K, Shaw S, Steinke K, et al. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019;47(W1):W81–W87.
- Park J, Park J, Jang S, et al. FTFD: an informatics pipeline supporting phylogenomic analysis of fungal transcription factors. Bioinformatics. 2008;24(7):1024–1025.
- Park J, Lee S, Choi J, et al. Fungal cytochrome P450 database. BMC Genomics. 2008;9(1):402.
- Bao W, Kojima KK, Kohany O. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob Dna. 2015;6:11.
- Kapitonov VV, Jurka J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat Rev Genet. 2008;9(5):411–412. Author reply 414.
- Smit AFA, Hubley R, Green P. RepeatMasker. [Internet]. 2015. Available from: http://repeatmasker.org.
- Chan PP, Lowe TM. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 2019;1962:1–14.
- Burge SW, Daub J, Eberhardt R, et al. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 2013;41(D1):D226–D232.
