Abstract
In Pleurotus sp., green mold, which is considered a major epidemic, is caused by several Trichoderma species. To develop a rapid molecular marker specific for Trichoderma spp. that potentially cause green mold, eleven Trichoderma species were collected from mushroom farms and the Korean Agricultural Culture Collection (KACC). A dominant fungal isolate from a green mold-infected substrate was identified as Trichoderma pleuroticola based on the sequences of its internal transcribed spacer (ITS) and translation elongation factor 1-α (tef1) genes. In artificial inoculation tests, all Trichoderma spp., including T. atroviride, T. cf. virens, T. citrinoviride, T. harzianum, T. koningii, T. longibrachiatum, T. pleurotum, and T. pleuroticola, showed pathogenicity to some extent, and the observed symptoms were soaked mycelia with a red-brown pigment and retarded mycelium regeneration. A molecular marker was developed for the rapid detection of wide range of Trichoderma spp. based on the DNA sequence alignment of the ITS1 and ITS2 regions of Trichoderma spp. The developed primer set detected only Trichoderma spp., and no cross reactivity with edible mushrooms was observed. The detection limits for the PCR assay of T. harzianum (KACC40558), T. pleurotum (KACC44537), and T. pleuroticola (CAF-TP3) were found to be 500, 50, and 5 fg, respectively, and the detection limit for the pathogen-to-host ratio was approximately 1:10,000 (wt/wt).
1. Introduction
Pleurotus eryngii (king oyster mushroom) is widely consumed in East Asia and has increased in popularity in Europe and North America [Citation1,Citation2] due to its remarkable flavor and high nutritional value [Citation3,Citation4]. Thus, the significance and production of this mushroom are increasing, and the main countries that produce this mushroom are China (1,360,000 M/T, 2015) (Korea Agriculture Trade Information, KATI, http://www.kati.net), Korea (49,136 M/T, 2018, Ministry of Agriculture, Food and Rural Affairs, http://www.mafra.go.kr/), and Japan (39,411 M/T, 2018, Forestry Agency of Japan, http://www.rinya.maff.go.jp/).
P. eryngii is cultivated using a polypropylene (PP) bottle system involving filling PP bottles with media, sterilization with steam, cooling, inoculation, spawn running, scraping old spawn, fruiting, and harvest [Citation5]. Many mushroom farms for P. eryngii production are well built with sanitation systems and automation [Citation6], but diseases and pests are still serious threats to high quality and stable production.
A wide range of Trichoderma species have been found to be associated with green mold in Pleurotus spp. Specifically, T. harzianum and T. aggressivum exhibit potent virulence to Pleurotus spp., as demonstrated by considerable yield losses and quality reductions [Citation7,Citation8]. T. atroviride, T. cf. virens, T. citrinoviride, T. koningii, T. longibrachiatum, T. pleurotum, and T. pleuroticola have also been reported as causal agents of Pleurotus [Citation8–11]. Additionally, T. pleurotum and T. pleuroticola have been found to suppress the growth of P. eryngii [Citation12] in vitro.
The genus Trichoderma is ubiquitous in various environments, including soil, forest, and root ecosystems [Citation13], and several Trichoderma species have even been isolated from a natural habitat and a fruiting body of Pleurotus [Citation10]. However, controlling the diseases caused by Trichoderma could be challenging due to the difficulty in distinguishing pathogenic mycelia from mushroom mycelia at the early growth stage because both are white. Moreover, Trichoderma spp. produce large numbers of conidia during asexual development [Citation13], which can be transferred throughout mushroom farms by ventilation and workers because these species are airborne and have a high ability to adhere to clothes [Citation14]. Monitoring the Trichoderma population in incubation rooms and scraping rooms is thus critical. Therefore, the development of an effective detection method for Trichoderma spp. in both mycelial cultures and environments is important for decreasing yield losses during mushroom cultivation.
In this study, we collected Trichoderma spp. from infected substrates and microorganism stock centers and investigated their pathogenicity. A molecular marker was then developed for the rapid and specific detection of Trichoderma spp.
2. Materials and methods
2.1. Fungal strains and growth conditions
A total of 11 Trichoderma strains were used, one was isolated in this work, and the others were from microorganisms stock center. Fungal isolates from substrate of bottles infected with green mold in P. eryngii cultivation farm in Jinju, Korea, were isolated by diluting and plating the sampled material on mushroom complete media (MCM; 0.2% peptone, 0.2% yeast extract, 2.0% glucose, 0.05% MgSO4·7H2O, 0.05% K2HPO4, and 0.046% KH2PO4). The plates were incubated at 25.0 °C until fungal mycelia appeared, and the mycelia were then transferred to fresh MCM medium for identification. Dominant and dark greenish fungi were selected for further experiments. Ten strains of Trichoderma spp. were provided by the Korean Agricultural Culture Collection (KACC), Jeonju, Korea (). P. eryngii, P. ostreatus, and Grifola frondosa were obtained from the Gyeongnam Agricultural Research and Extension Services, Jinju, Korea (), and used as controls in the pathogenicity test and PCR assays. All fungal isolates and strains were maintained on MCM in the dark at 25 °C with periodic transfers.
Table 1. List of Trichoderma spp. and mushrooms used in this study.
2.2. Genomic DNA extraction and sequencing of ITS and TEF-1α regions
The fungal genomic DNA (gDNA) was extracted from lyophilized mycelia using a GenEx Plant plus! Kit (GeneAll, Seoul, Korea) as described previously [Citation15]. The internal transcribed spacer 1 (ITS1), 5.8S rRNA, and ITS2 regions were amplified by PCR using the ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) primers [Citation16]. The tef1 gene was amplified using the translation elongation factor 728 (TEF728) (GCCATCCTTGGAGATACCAGC) and TEF1 (CATCGAGAAGTTCGAGAAGG) primers [Citation17] in a 20-µl reaction mixture as described previously [Citation18]. The PCR products were analyzed in a 1.0% agarose gel (Promega, Madison, WI, USA) containing SafeView Classic (Intron Biotechnology, Seongnam, Korea). The specific bands were then cut out and sequenced [Citation19]. For each reaction, at least three different amplified genes were bidirectionally sequenced. The sequence that corresponds to tef1 gene was identified by an alignment with identical genes from GenBank. The sequences obtained in this study have been deposited in GenBank under the accession number MK611085 (ITS) and MT219315 (tef1). The ITS and tef1 sequences were identified using TrichOKEY 2 [Citation20] and TrichoBLAST [Citation21] in combination with the NCBI-BLAST database.
2.3. Pathogenicity test
Trichoderma spp. were grown on MCM at 25 °C for 14 days, and the conidia were harvested with distilled water. The suspensions were filtered through a nylon membrane (40 µm; Millipore, Billerica, MA, USA) and resuspended to about 1.4 × 108 conidia per ml, as determined by direct counting using a hemocytometer.
The fructification of P. eryngii KNT2312 was analyzed as reported previously [Citation22]. At the end of the mycelial run (35 days at 25 °C), the old spawn and top layer of substrate (∼1 cm) were removed by scraping to induce fruiting. Subsequently, 1 ml of Trichoderma solution containing approximately 1.4 × 108 conidia was inoculated onto the substrate surface in each PP bottle, and the cultures were placed in a room that was maintained at 15 °C with 95% relative humidity and cool-white fluorescence light (200 Lux). The control culture was inoculated with distilled water. The fruiting bodies were harvested before the pileus had opened completely to determine various phenotypic traits, including the yield, pileus, and stipe, using calipers (Mitutoyo, Tokyo, Japan) and a balance as described previously [Citation22]. The earliness (days to harvest after removing old medium) was also measured. Four repetitions were performed for each of the 11 Trichoderma spp. and the control.
The statistical significance of the yield and earliness in the presence of different Trichoderma spp. was determined by comparing the group means through analysis of variance (ANOVA) followed by Duncan’s multiple-range test (DMRT). Statistically significant differences were determined at p < 0.05. The data analyses were performed using R open-source software [Citation23].
2.4. Primer design for potential causative Trichoderma spp
The sequences of the ITS regions of Trichoderma spp. with those of edible mushrooms were compared using Clustal Omega software to identify specific sequences for Trichoderma spp. [Citation24]. In addition to the ITS sequence of T. pleuroticola (CAF-TP3), additional ITS sequences were retrieved from GenBank: T. pleuroticola, EU918140; T. pleuroti, NR_134421; T. pleurotum, EU280069; T. aggressivum, AY154947; T. aureoviride, AF194007; T. harzianum, EU918151; P. eryngii, FJ90770; P. ostreatus, FN391585; Agaricus bisporus, AJ409229; Flammulina velutipes, KJ999151; and Lentinula edodes, AF079572. The specific sequences were used to design specific primers targeting the consensus sequences in the ITS1 and ITS2 regions of Trichoderma with the aim of amplifying only those of Trichoderma spp. and not those of edible mushrooms. The developed primer sequences were TDP-F: 5′-CGAGTTTACAACTCCCAAA-3′ and TDP-R: 5′-GAAAGTTGGGTGTTTAACG-3′. The primer set was tested by in silico PCR on ITS sequences of Apergillus and Penicillium from GenBank (NR_077154.1, NR_077145.1, NR_111348.1, NR_111041.1, NR_151784.1) for selectivity using FastPCR software [Citation25].
2.5. PCR conditions, sensitivity, and specificity of PCR assays
PCR was performed using Ex Taq (Takara, Kyoto, Japan) and the following conditions: initial denaturation at 98 °C for 30 s, 35 cycles of denaturation at 98 °C for 10 s, annealing at 60 °C for 30 s, and extension at 72 °C for 1 min, and one cycle of final extension at 72 °C for 7 min. The PCR products were resolved on a 1% agarose gel containing SafeView Classic. To assess the diagnostic sensitivity, 10-fold serial dilutions of T. harzianum (KACC40558), T. pleurotum (KACC44537), and T. pleuroticola (CAF-TP3) gDNA, ranging from 5 ng to 5 ag per 20-μl PCR, were prepared. To evaluate the specificity of the PCR, serially diluted mixtures (10-fold) of host:pathogen gDNA ranging from 1:10 to 1:100,000 (wt/wt) were prepared: P. eryngii KNR2312 gDNA with T. harzianum (KACC40558), T. pleurotum (KACC44537) or T. pleuroticola (CAF-TP3) gDNA. The PCRs were performed under the same aforementioned conditions.
3. Results
3.1. Identification of greenish fungus from substrate infected with green mold
The dominant fungi appeared to grow rapidly on MCM and exhibited dark green mycelia. Isolates (CAF-TP3) were selected based on their morphology compared with the typical morphology of Trichoderma spp. The ITS1 and ITS2 regions were amplified by PCR using the universal primers ITS1 and ITS4. The sequence of the amplicon was 639 bp and shared 98.6% identity with other T. pleuroticola in NCBI. The TrichOKEY v.2.0 identification profile showed three barcode sequences in ITS1 and ITS2: GATCTCTG, GTTTTTTTATAATCT, and CCCCTCGTGGG. These barcode sequences identified the isolate as T. pleuroticola with high reliability in the TrichOKEY.
PCR using the TEF728 and TEF1 primer set amplified a single band of 2237 bp. The sequence corresponding to tef1 gene was estimated to be 613 bp long. As determined with TrichoMARK, the sequence of the tef1 gene of the CAF-TP3 contained two phylogenetic markers, namely, 301 bp of tef_int4 (large) and 86 bp of tef1_int5 (short), which share 99.3% and 100% with other T. pleuroticola in NCBI, respectively. In addition, 3 of 12 known tef anchors, EF1-986r (GGCAAGGGTT), tef1fw (GTGAGCGTG), and EF2 (ACTGGTAC), were found in the tef1 sequence.
3.2. Pathogenicity of Trichoderma spp. to P. eryngii
Trichoderma strains used in this study need to be ensure their pathogenicity to P. eryngii because they were isolated from various sources and their severity of green mold was obscure, even though all were associated with green mold. The appearance and development of the fruiting body and green mold symptoms were monitored daily. At the initial stage of fruiting body development, mycelia covered the substrate and formed condensed mycelia that became primordia. However, the Trichoderma-inoculated substrates were not covered or were slowly covered with mycelia. The grown mycelia of P. eryngii produced a reddish-brown pigment on all substrates after inoculation with Trichoderma spp. The main symptoms were soaked mycelia and retarded mycelial regeneration, which resulted in lack of fruiting, yield loss, and late harvest. Primordia did not appear on the substrate inoculated with T. pleurotum KACC44537, whereas on the substrate inoculated with T. cf. virens KACC40783, primordia appeared in a few areas (). Small fruiting bodies and late harvests were observed with the remaining Trichoderma species ()). All Trichoderma species caused yield loss, ranging from 20.5% to 99.4% compared with that obtained with the control, and late harvest (). Prolonged earliness was observed on all the substrates treated with Trichoderma species, ranging from 0.3 to 6.0 days compared with that obtained with the control. T. harzianum (KACC40784) did not cause significant yield loss (238.6 g vs. 240.0 g) but led to late harvest (20.8 vs. 17.0 days) (). The statistical analysis showed that the yield and earliness were not significantly different between the treatments. However, the yields obtained in the three Trichoderma-treated plots (T. pleurotum KACC44537, T. atroviride KACC40774, and T. cf. virens KACC40783) were significantly lower than that of the control (p < 0.05), and the measured earliness in six Trichoderma-treated plots was statistically significantly later than that of the control. Moreover, the quality and hardness of the fruiting bodies were lower than those of the control. After Trichoderma infection, a reddish brown lesion was found inside the base stipe, and the shelf life of the fruiting body after infection was shorter compared with that of the control (data not shown).
Figure 1. Symptoms of green mold in the fruiting body of P. eryngii after artificial inoculation with Trichoderma spp. (A) Control (distilled water); (B) T. harzianum (KACC40558); (C) T. longibrachiatum (KACC40563); (D) T. atroviride (KACC0774); (E) T. koningii (KACC40779); (F) T. cf. virens (KACC40783); (G) T. harzianum (KACC40784); (H) T. pleuroticola (KACC44535); (I) T. pleuroticola (KACC44536); (J) T. pleurotum (KACC44537); (K) T. citrinoviride (KACC44703); and (L) T. pleuroticola (CAF-TP3). Each inoculation was performed with 1.4 × 108 conidia. Picture were taken 18 days after inoculation.
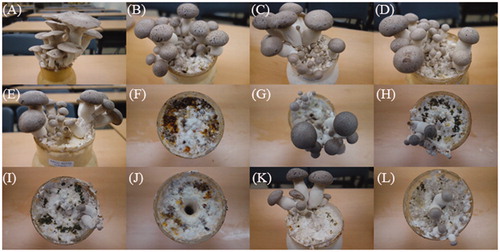
Table 2. Phenotypic characteristics of Pleurotus eryngii artificially infected with Trichoderma spp.
3.3. Detection performance, sensitivity, and specificity of the developed primer set
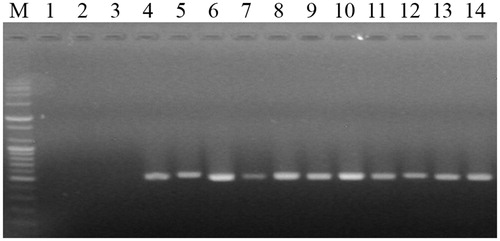
A sequence alignment analysis showed the ITS regions of Trichoderma spp. and edible mushrooms (). Several forward and reverse primer sets were designed based on the consensus sequences flanking the 18S rRNA and 28S rRNA sequences (). The developed PCR sets were tested using Trichoderma spp. and several edible mushroom gDNA extracts. A single amplicon was observed for each Trichoderma sp. at the expected size (∼530 bp) with the TDP-F and TDP-R primer combination, and no cross-reactivity could be observed with the edible mushrooms (). In addition, the primer set showed no amplicon with genome sequences of Aspergillus and Penicillum in in silico PCR (data not shown).
Figure 2. Sequence comparison of the ITS1 and ITS2 regions of Trichoderma spp. and edible mushrooms. The conserved regions of Trichoderma spp. are shaded in gray in the sequences corresponding to the specific primer set TDP-F and TDP-R.
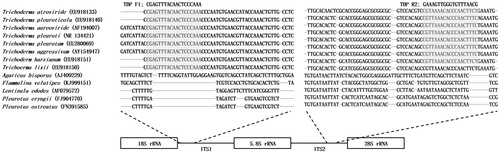
Figure 3. DNA fragments amplified by PCR using the TDP-F and TDP-R primer set specific for Trichoderma spp. M, 100-bp plus DNA ladder (Bioneer, Daejeon, Korea); lane 1, Pleurotus eryngii; lane 2, P. ostreatus; lane 3, Grifola frondosa; lane 4, T. harzianum (KACC40558); lane 5, T. longibrachiatum (KACC40563); lane 6, T. atroviride (KACC40774); lane 7, T. koningii (KACC40779); lane 8, T. cf. virens (KACC40783); lane 9, T. harzianum (KACC40784); lane 10, T. pleuroticola (KACC44535); lane 11, T. pleuroticola (KACC44536); lane 12, T. pleurotum (KACC44537); lane 13, T. citrinoviride (KACC44703); and lane 14, T. pleuroticola (CAF-TP3).
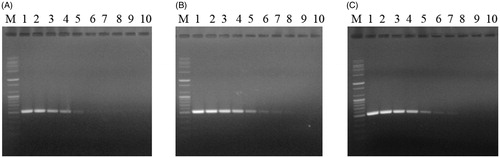
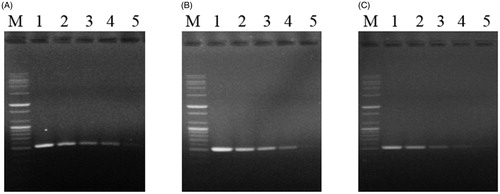
The application of the TDP-F and TDP-R primer set to the 10-fold dilution series of T. harzianum (KACC40558), T. pleurotum (KACC44537), and T. pleuroticola (CAF-TP3) gDNA consistently revealed detection limits of 50 fg, 500 ag, and 5 fg, respectively, after 35 cycles of amplification ().
Figure 4. Sensitivity of the TDP-F and TDP-R primer set for pathogen DNA extracts. DNA bands amplified by PCR with the TDP-F and TDP-R primer set from 10-fold serial dilutions of gDNA from 5 ng to 5 ag of (A) T. harzianum (KACC40558); (B) T. pleurotum (KACC44537); or (C) T. pleuroticola (CAF-TP3). M, 100-bp plus DNA ladder (Bioneer, Daejeon, Korea); lane 1, 5 ng; lane 2, 500 pg; lane 3, 50 pg; lane 4; 5 pg; lane 5, 500 fg; lane 6, 50 fg; lane 7, 5 fg; lane 8, 500 ag; lane 9, 50 ag; and lane 10, 5 ag.
PCR using the developed primer set could amplify a single amplicon at a pathogen:P. eryngii ratio as low as 1:10,000 for all tested Trichoderma spp. (). Weak amplicons were observed at a ratio of 1:100,000, but the band was likely unreliable.
Figure 5. Specificity of the TDP-F and TDP-R primer set for host–pathogen DNA mixtures. DNA bands amplified by PCR with the TDP-F and TDP-R primer set from serially diluted mixtures (10-fold) of P. eryngii KNR2312 gDNA with gDNA from (A) T. harzianum (KACC40558); (B) T. pleurotum (KACC44537); or (C) T. pleuroticola (CAF-TP3) prepared at ratios ranging from 1:10 to 1:100,000 (wt/wt). M, 100-bp plus DNA ladder (Bioneer, Daejeon, Korea); lane 1, 1:10 (wt/wt) ratio of pathogen:P. eryngii; lane 2, 1:100 (wt/wt) ratio of pathogen:P. eryngii; lane 3, 1:1000 (wt/wt) ratio of pathogen:P. eryngii; lane 4; 1:10,000 (wt/wt) ratio of pathogen:P. eryngii; and lane 5, 1:100,000 (wt/wt) ratio of pathogen:P. eryngii.
4. Discussion
In this study, we isolated a dark green fungus from a substrate of P. eryngii infected with green mold and identified it as T. pleuroticola. Three barcode sequences from ITS sequences and two phylogenetic markers from the tef1 gene, tef_int4 (large) and tef1_int5 (short), confirmed that the isolate was T. pleuroticola. T. harzianum and its biotypes are known as severe pathogens to A. bisporus [Citation26], whereas T. pleurotum and T. pleuroticola are frequently found in Pleurotus mushroom farms [Citation9–11] but not in P. eryngii. To our knowledge, this study describes the first isolation of T. pleuroticola from a substrate of P. eryngii.
All Trichoderma species showed pathogenicity, as demonstrated by a lack of fruiting bodies and soaked lesions. Interestingly, T. cf. virens and T. pleurotum, which are not known for their pathogenic severity, even though T. cf. virens was previously found to inhibit the mycelial growth of P. eryngii [Citation27], caused no or very less fruiting body formation (). T. pleurotum and T. pleuroticola have also been reported to reduce the mycelial growth of P. eryngii [Citation12]. In our study, pathogenicity was not consistent within species (e.g., T. pleuroticola KACC44535 and KACC44536 showed relatively moderate and severe pathogenicity, respectively). These results reinforce the notion that T. harzianum and T. aggressivum are closely related but exhibit a broad pathogenicity range [Citation28].
Earliness is also economically important because this measure is related to fast turnover in a cultivation room. Although no significant difference in earliness was found between the control and six treated plots (50%), all Trichoderma spp. were found to delay the harvest, which might have been caused by suppression of primordia formation, and the observation of soaked mycelia indicate that the mechanism underlying this suppression might involve lysis of the mycelium ().
We developed a rapid and accurate detection method for 11 Trichoderma spp. that involves a single primer set. The molecular markers TDP-F and TDP-R can detect the mycelium of Trichoderma not only independently () but also in a mixture of Trichoderma and edible mushrooms (). Although detection methods for T. harzianum, T. pleurotum, and T. pleuroticola have been developed in previous studies [Citation10,Citation29,Citation30], the detection ranges of these previously developed methods are limited. Because the 11 Trichoderma spp. tested in our study were found to exhibit pathogenicity to P. eryngii, the range of detection is important for the control of green mold. In in silico PCR, Penicillium and Aspergillus, another green molds with weak pathogenicity, were not detected with the developed primer set. We could not exclude the possibility that the developed marker could be positive on other Trichoderma species besides the ones tested in this study. It would not a big problem, because the nature of Trichoderma is parasite of other fungi [Citation13], thus they might be antagonistic effect on edible mushrooms.
In our study, the detection limits for Trichoderma spp. ranged from 500 ag to 5 fg (). These sensitivities are similar to those of nested PCR assays with specific primers targeting Trichoderma spp. [Citation29], but the method developed in our study has enhanced usability because it utilizes the standard PCR protocol. The developed primer set was able to detect a 1:10,000 (wt/wt) ratio of Trichoderma spp.:P. eryngii mycelium (), which reveals the detection effectiveness of the developed molecular markers during the initial growth phase of Trichoderma spp. on the mushroom substrate. This specificity is very important because the ratio of Trichoderma mycelium:P. eryngii might be very low at the early stage of spawn running. This information will help farms reduce contamination in liquid spawn systems, which has recently increased in Korea [Citation31,Citation32]. The liquid spawn is applied with air pressure through a membrane filter, which might increase the risk of exposure to substantial contamination from airborne Trichoderma spores.
The control of Trichoderma can be challenging due to its features, habitats, optimal temperature, aggressive growth rate, and airborne conidia [Citation13]. In addition, the substrate is very vulnerable after the scraping of old mycelia, which act as a barrier. Before new mycelia can grow and cover the surface of the substrate, the scraped substrate might be exposed to airborne pathogens such as Trichoderma. Thus, the detection and removal of pathogens at the early stage and reducing their population in mushroom farms will be the best strategy. Consequently, the molecular markers TDP-F and TDP-R will lead to better management of commercial mushroom production and prevent green mold caused by Trichoderma spp.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Mleczek M, Siwulski M, Rzymski P, et al. Cultivation of mushrooms for production of food biofortified with lithium. Eur Food Res Technol. 2017;243(6):1097–1104.
- Estrada AR, Royse DJ. Yield, size and bacterial blotch resistance of Pleurotus eryngii grown on cottonseed hulls/oak sawdust supplemented with manganese, copper and whole ground soybean. Bioresour Technol. 2007;98(10):1898–1906.
- Jin X, Wang Q, Yang X, et al. Chemical characterisation and hypolipidaemic effects of two purified Pleurotus eryngii polysaccharides. Int J Food Sci Technol. 2018;53(10):2298–2307.
- Ma G, Kimatu BM, Zhao L, et al. Impacts of dietary Pleurotus eryngii polysaccharide on nutrient digestion, metabolism, and immune response of the small intestine and colon – an iTRAQ-based proteomic analysis. Proteomics. 2018;18(7):1700443.
- Lee SH, Kim MK, Ryu JS, et al. Characteristics of a new Pleurotus eryngii cultivar, Aeryni 6. J Mushroom. 2018;16(1):16–21.
- Jang MJ, Park Y, Kim JH. Properties of disease occurrence by season for cultivation facilities of oyster mushroom. J Mushroom Sci. 2019;17(3):93–98.
- Sobiralski K, Siwulski M, Gorski R, et al. Impact of Trichoderma aggressivum f. europaeum isolates on yielding and morphological features of Pleurotus eryngii. Pytopathologia. 2010;56:17–25.
- Hatvani L, Antal L, Manczinger L, et al. Green mold diseases of Agaricus and Pleurotus spp. are caused by related but phylogenetically different Trichoderma species. Phytopathology. 2007;97(4):532–537.
- Komoñ-Zelazowska M, Bissett J, Zafari D, et al. Genetically closely related but phenotypically divergent Trichoderma species cause green mold disease in oyster mushroom farms worldwide. Appl Environ Microbiol. 2007;73(22):7415–7426.
- Kredics L, Kocsubé S, Nagy L, et al. Molecular identification of Trichoderma species associated with Pleurotus ostreatus and natural substrates of the oyster mushroom. FEMS Microbiol Lett. 2009;300(1):58–67.
- Park MS, Bae KS, Yu SH. The new species of Trichoderma associated with green mold of oyster mushroom cultivation in Korea. Mycobiology. 2006;34(3):11–13.
- Sobieralski K, Siwulski M, Komon-Żelazowska M, et al. Evaluation of the growth of Trichoderma pleurotum and Trichoderma pleuroticola isolates and their biotic interaction with Pleurotus sp. J Plant Protect Res. 2012;52(2):235–239.
- Harman GE, Howell CR, Viterbo A, et al. Trichoderma species-opportunistic, avirulent plant symbionts. Nat Rev Microbiol. 2004;2(1):43–56.
- Wellings CR, McIntosh RA, Walker J. Puccinia striiformis f. sp. tritici in Eastern Australia –possible means of entry and implications for plant quarantine. Plant Pathol. 1987;36(3):239–241.
- Park B, Ha BS, Lee SH, et al. Variable-number tandem repeat loci-discriminating Pleurotus ostreatus cultivars. Mycoscience. 2019;60(2):132–135.
- White TJ, Brunes T, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M, Gelfand D, Sninsky J, et al., editors. PCR protocols: a guide to methods and applications. Cambridge, MA: Academic Press; 1990. p. 315–322.
- Dees PM, Ghiorse WC. Microbial diversity in hot synthetic compost as revealed by PCR-amplified rRNA sequences from cultivated isolates and extracted DNA. FEMS Microbiol Ecol. 2001;35(2):207–216.
- Lee SH, Ali A, Ha B, et al. Development of a molecular marker linked to the A4 locus and the structure of HD genes in Pleurotus eryngii. Mycobiology. 2019;47(2):200–206.
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74(12):5463–5467.
- Druzhinina IS, Kopchinskiy AG, Komoń M, et al. An oligonucleotide barcode for species identification in Trichoderma and Hypocrea. Fungal Genet Biol. 2005;42(10):813–828.
- Kopchinskiy A, Komoń M, Kubicek CP, et al. TrichoBLAST: a multilocus database for Trichoderma and Hypocrea identifications. Mycol Res. 2005;109(Part 6):658–660.
- Ryu JS, Kim MK, Im CH, et al. Development of cultivation media for extending the shelf-life and improving yield of king oyster mushrooms (Pleurotus eryngii). Sci Hortic. 2015;193:121–126.
- Team RDC. R: a language and environment for statistical computing. Vienna, Austria: R foundation for statistical computing; 2013. Available from: http://www.R-project.org.
- Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. 2002;Chapter 2:Unit 2.3.
- Kalendar R, Lee D, Schulman AH. FastPCR software for PCR, in silico PCR, and oligonucleotide assembly and analysis. In: Valla S, Lale R, editors. DNA cloning and assembly methods, Methods in Molecular Biology. Totowa, NJ: Humana Press; 2014. p. 271–302.
- Williams J, Clarkson JM, Mills PR, et al. Saprotrophic and mycoparasitic components of aggressiveness of Trichoderma harzianum groups toward the commercial mushroom Agaricus bisporus. Appl Environ Microbiol. 2003;69(7):4192–4199.
- Choi I-Y, Joung G-T, Ryu J, et al. Physiological characteristics of green mold (Trichoderma spp.) isolated from Oyster Mushroom (Pleurotus spp). Mycobiology. 2003;31(3):139–144.
- Samuels GJ, Dodd SL, Gams W, et al. Trichoderma species associated with the green mold epidemic of commercially grown Agaricus bisporus. Mycologia. 2002;94(1):146–170.
- Miyazaki K, Tsuchiya Y, Okuda T. Specific PCR assays for the detection of Trichoderma harzianum causing green mold disease during mushroom cultivation. Mycoscience. 2009;50(2):94–99.
- Kim SW, Kim S, Lee HJ, et al. Isolation of fungal pathogens to an edible mushroom, Pleurotus eryngii, and development of specific ITS primers. Mycobiology. 2013;41(4):252–255.
- Janpoor J, Pourianfar HR, Shahtahmasebi S. Study on effect of culture medium and growth conditions on liquid spawn king oyster mushroom (Pleurotus eryngii). Int J Farm Allied Sci. 2017;6(6):154–156.
- Lee SJ, Kim HH, Kim SH, et al. Culture conditions of liquid spawn and the growth characteristics of Pleurotus ostreatus. J Mushroom. 2018;16(3):162–170.
