Abstract
Plum pocket caused by the dimorphic ascomycetous fungi, Taphrina spp., results in unsightly malformations and crop loss. In 2016, Japanese plums (Prunus salicina Lindl.) with plum pocket symptoms were found in Gimcheon. Three isolates were collected from symptomatic P. salicina fruits and identified as Taphrina deformans based on morphological characteristics and molecular sequence analysis of including internal transcribed space (ITS) and the mitochondrial small ribosomal subunit (SSU) regions of the three isolates. Pathogenicity test on plum fruits confirmed that, the present T. deformans isolates are causal agent of plum pocket. To the best of our knowledge, this is the first report of plum pocket caused by T. deformans in South Korea.
Keywords:
The Japanese plum (Prunus salicina Lindl.) is considered to have originated in the Yangtze River basin and has been domesticated in Japan [Citation1,Citation2]. It is grown across a wide range of climatic zones worldwide [Citation3]. The People’s Republic of China, European Union, Romania, Serbia, USA, and Turkey are the major plum growing countries [Citation4]. Japanese plum is also cultivated across South Korea. The Japanese plum takes the sixth place in terms of deciduous fruit production in South Korea, following apple, persimmon, peach, grape, and pear [Citation5,Citation6]. According to the estimated data from FAO [Citation4], the total production of the Japanese plum in The Republic of Korea was 74,246 tons in 2016 growing season. The Japanese plum is mostly eaten in the fresh form and occasionally in the dried form worldwide. The Japanese plum fruits are rich in organic acids, carbohydrates, flavonoids, and anthocyanins and are effective in preventing various diseases [Citation7].
Among the fungal diseases, plum pocket has most frequently occurred in South Korea over the recent years. It has been reported that outbreak of this disease results in a significant yield loss (>30%) if regular spray programs are not implemented [Citation8]. Accurate identification of causal agent is very important to develop sustainable and eco-friendly control measures of this disease. Taphrina species including Taphrina pruni (Fuck.) Tul and T. communis (Sadeb.) are responsible for plum pocket [Citation9–11]. Differentiation of a majority of Taphrina species has been made on the basis of the morphological and physiological traits of their hosts. Distinguishing Taphrina species based on host specificity and morphological characteristics has not yet been thoroughly validated [Citation9,Citation12,Citation13]. In this case, molecular methods give a good overview of Taphrina species differentiation or identification [Citation10]. Because of the scarcity of molecular studies and available cultures, there has been little progress in the morphological and molecular characterization of causal agents for plum pocket in South Korea.
The main objectives of this study were to identify the causal agent of plum pocket based on morphological characteristics and molecular analysis.
In 2016, plum pockets were observed in a majority of plum orchards in Gimcheon, Korea. The disease caused distorted and enlarged fruits with a spongy or hollow center (). The deformed fruits became dark brown in color and eventually fell from trees. For morphological characterization of perfect stages, compact layer of asci was excised from deformed plum fruits and subsequently squashed in lactophenol-acid cotton blue and examined at a magnification of 400×. Sizes of intake ascus and ascospores were measured using a stage micrometer under an Olympus BX43 microscope (Olympus, Tokyo, Japan) at 400× magnification. T. deformans produces a white layer of asci on enlarged fruits of P. salicina (). The asci are cylindrical to clavate cylindrical with a dimension of 21.58–50.03 × 6.98–14.78 μm (n = 30) (mean: 41.38 × 10.95 μm); they are round at the apex (). The ascospores are oval or ovoid to spherical and measure 5.72–7.90 × 4.50–7.20 μm in size (n = 30) (mean: 7.08 × 5.64 μm) ().
Figure 1. Plum pocket of plum and sexual stage of Taphrina deformans. (A) Deformed plum fruits; (B) Deformed fruits inoculated with T. deformans yeast cells; (C) Asci; (D) Ascospores.

The yeast strains of Taphrina spp. (T. deformans) described in this study were isolated from infected fruits using spore-fall methods [Citation14]. Wet infected fruit tissue with mature asci was attached to the lids of a petri dish, thereby allowing the ascospores to be ejected from asci onto the potato dextrose agar (PDA) plates supplemented with 0.5 g L−1 streptomycin sulfate (PDAS). Plates were incubated at 25 °C in the dark until yeast colonies began emerging. The newly emerged yeast colonies were transferred to PDA plates to understand colony morphology. By following this procedure, 10 strains were isolated and identified as Taphrina spp. based on colony color and yeast-like colony growth. A subsample of three stains (NOOT1, NOOT2, and NOOT3) was used for morphological and molecular characterization.
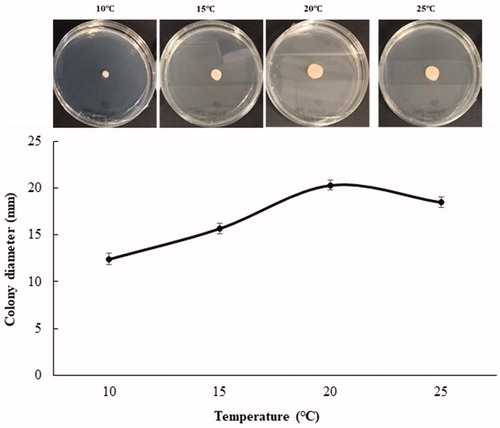
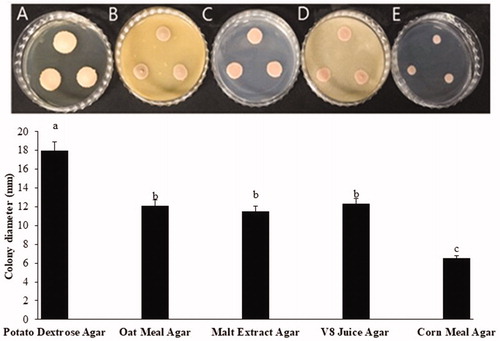
Strain NOOT1 was selected for morphological characterization of yeast stage of T. deformans. To examine the size and shape of the yeast cell, the temporary slide was prepared. Culture was taken from the edge of a colony on a glass slide and one drop of lactoglycerol solution was added to it. Subsequently, the slide was tested under a microscope and the size of yeast cells was measured in the same manner as the measurements were taken for asci and ascospores. The yeast cultures grow slowly on PDA. Twenty-one-day-old colonies ranged from pale pink and pinkish cream to salmon pink coloration on both sides; they were butyrous with a smooth lustrous surface (). NOOT1 cells were ovoid to ellipsoidal, with a measurement of 4.42–8.6 × 2.8–5.4 μm (n = 50) (mean: 6.2 × 3.8) in size and occurred singly or with buds (). Occasionally, yeast cells of the strain NOOT1 clung together in chains and produced a pseudo-mycelium (resembling small true mycelia) (). Growth rate on different media including PDA, cornmeal agar (CMA), oatmeal agar (OA), malt extract agar (MA), and V8 juice agar (V8) was determined. NOOT1 was cultured on each media and incubated at 20 °C under16:6 h of light: dark condition. To determine the growth rate under different temperatures, NOOT1 was cultured on PDA and incubated at 10, 15, 20, and 25 °C under 16:6 h of light: dark conditions. Growth rate was measured during each week. The growth rates of culture during each treatment were compared using the least significant difference (LSD) tests (α = 0.05) as well as by employing JMP Pro 10.0.0 software. Media significantly (p<.001) affected colony growth of T. deformans (). Colonies grew significantly faster on PDA. OA, MA, and V8 also favored colony growth. CMA was not good for colony growth (). Colonies grew actively at temperatures from 10 to 25 °C. Radial growth rate increased until a temperature of 20 °C, and then decreased with increase in temperature (). The morphological characteristics including asci, ascospores, and yeast cell size of T. deformans isolates from plum were consistent with those mentioned in previous reports for Taphrina spp. [Citation9,Citation14,Citation15].
Figure 2. Yeast stage of Taphrina deformans on potato dextrose agar plates. (A) Obverse colony; (B) Reverse side of the colony; (C) Yeast cells; (D) Budding yeast cells and pseudo mycelium.
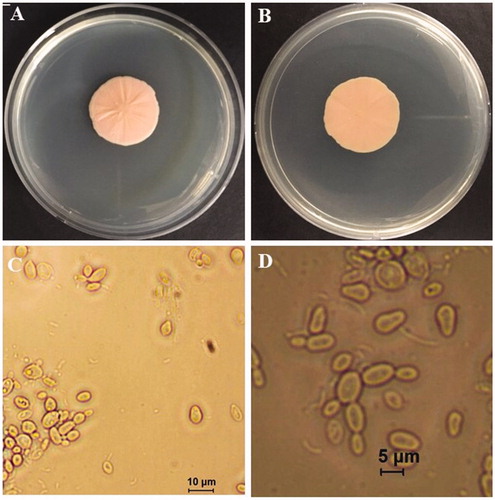
Figure 3. Effect of culture media on colony growth of Taphrina deformans at 20. (A) Potato dextrose agar; (B) Oat meal agar; (C) Malt extract agar; (D) V8 juice agar; (E) Corn meal agar. Different letter on bar indicate that mean are significantly different (p<.05) according to Fisher’s LSD test.
The total genomic DNA were extracted from 20-day-old cultures of NOOT1, NOOT2, and NOOT3 isolates using a HiGeneTM Genomic DNA Prep Kit (BIOFACT, Daejeon, Korea) by following the manufacturer’s instructions. The isolated DNA from fungal isolates used in this study was used as a template for amplification by polymerase chain reaction (PCR). The primer sets ITS1F and ITS4 [Citation16,Citation17] and SSU1 and SSU5 [Citation9] were used for the amplification of the ITS-1, ITS-2, and 5.8 S gene regions of the ITS region of a ribosomal DNA and portion of the gene for the mitochondrial small rRNA subunit (SSU), respectively. The PCR reactions were run in SimpliAmp Thermal Cycler (Thermo Fisher Scientific, Waltham, MA) maintaining conditions described by Petrydesová et al. [Citation15]. All the resulting PCR products were purified using HiGeneTM PCR Purification Kit (BIOFACT) and sequenced directly using their respective primers at Macrogen, Inc. (Seoul, Korea). The generated DNA sequences were analyzed, and consensus sequences were computed using SeqMan version 7.1 from the Lasergene package (DNASTAR, Inc. Madison, WI). The resulting consensus sequences were deposited in GenBank (accession numbers LC312418- LC312420, LC312651- LC312653).
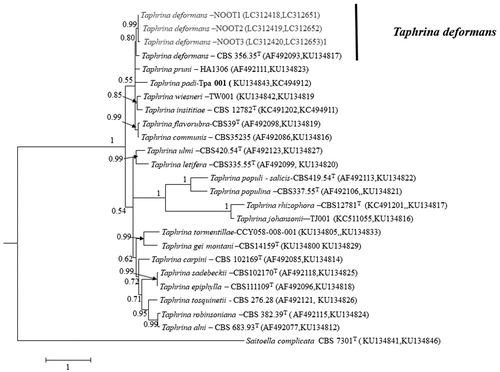
For phylogenetic analysis, sequences of ITS and SSU of previously identified Taphrina spp. [Citation9,Citation15] were retrieved from GenBank. Multiple sequence alignments of the present isolates and those retrieved from GenBank were prepared using MEGA version 6.0 [Citation18] which employed default muscle multiple sequence alignment programs. The aligned sequences of ITS and SSU were concatenated with Mesquite version 2.75 [Citation19]. Bayesian phylogenetic analysis was performed using MrBayes version 3.2.10 [Citation20]. Resulting tree was viewed in FigTree version 1. 3.1 [Citation21]. Posterior probability was used to evaluate reliability of each node. The 50% majority-rule consensus tree revealed that the fungal isolates of this study were clustered together in a sister clade with the T. demormans strain CBS 356.35 ().
Figure 5. Bayesian phylogeny (BI) based on a 50% majority rule consensus tree showing phylogenetic relationships based on the combined ITS and SSU DNA sequence alignment among the present isolates and Taphrina spp. The BI posterior probability (>0.5) are given at the nodes. The scale bar shows the number of substitutions per site. T, ex type strain.
A pathogenicity test was conducted in a commercial plum orchard near to the Kyungpook national university. The orchard was known to be plum pocket free in 2016. Six small twigs of 12-year-old P. salicina tree per isolate were selected. Yeast cell suspension of 1 × 106 CFU mL−1 was prepared from a 10-d-old culture and subsequently grown on PDA at 20 °C in the dark. On March 25 2017, the selected twigs were disinfected by spraying 70% ethanol. After drying, the twigs with unopened flower buds were inoculated by spraying the conidial suspension and were covered in a plastic bag. The twigs that received only distilled water spray served as the control. Disease incidence was estimated as the percentage of deformed fruits per twig. The yeast stage of the T. deformans isolates of this study was pathogenic to plum. The enlarged fruits with tongue-like outgrowths were observed on inoculated shoots with the yeast cell suspension of present isolates strain cells ().
Phylogenetic and couple with morphological analysis of this study revealed that the causal agent of plum pocket was T. deformans. Koch’s postulates were fulfilled by inoculating yeast cell suspensions of the obtained fungal isolates, on an attached, unopened flower bud; therefore, this study successfully confirmed that T. deformans was the causal agent of plum pocket disease. T. deformans is well known causal agent of peach leaf curl disease [Citation14,Citation15]. However, there is no report on plum pocket caused by T. deformans and to the best of our knowledge, this is the first report. Finding of this study will help to develop sustainable control measure of plum pocket in South Korea.
Acknowledgments
We would like to thank all lambaste, who helped us to run experiments successfully.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Faust M, Surányi D. Origin and dissemination of plums. Hort Rev. 1999;23:179–231.
- Fanning KJ, Topp B, Russell D, et al. Japanese plums (Prunus salicina Lindl.) and phytochemicals-breeding, horticultural practice, postharvest storage, processing and bioactivity. J Sci Food Agric. 2014;94(11):2137–2147.
- Hansmann CF, Combrink JC. Plums and related fruits. In: Caballero B, editor. Encyclopedia of food sciences and nutrition. 2nd ed. Cambridge (MA): Academic Press; 2003. p. 4606–4610.
- FAO. Food and agriculture organization of the United Nations. 2016. Available from: http://www.fao.org/faostat/en
- KOSIS. Korean statistical information service. 2016. Available from: http://kosis.kr
- Kwon JH, Nam EY, Jun JH, et al. Asian plum diversity based on phenotypic traits in republic of Korea. Kor J Plant Res. 2018;31:254–267.
- Bae H, Yun SK, Jun JH, et al. Assessment of organic acid and sugar composition in apricot, plumcot, plum, and peach during fruit development. J Appl Bot Food Qual. 2014;87:24–29.
- Behrendt C, Floyd C. Plum pockets: yard and garden brief. Minneapolis (MN): University of Minnesota, Extension Service; 1999. Available from: http://www.extension.umn.edu/yardandgarden/ygbriefs/p234plumpockets,html
- Rodrigues MG, Fonseca A. Molecular systematics of the dimorphic ascomycete genus Taphrina. Int J Syst Evol Microbiol. 2003;53(Pt 2):607–616.
- Prillinger H, BacigáLová K, Lopandic K, et al. Taphrina padi in Bayern und der Slowakei. Hoppea Denkschr Regensb Bot Ges. 2000;61:275–294.
- Ioana Jr M, Mitre V, Buta E, et al. Reaction of some plum cultivars to natural infection with Taphrina pruni (Fuck.) Tul. Fusicladium pruni ducomet and Tranzschelia pruni-spinosae person dietel. Agricultura. 2015;93:1–2.
- Mix AJ. A monograph of the genus Taphrina. Univ Kans Sci Bull. 1949;33:3–167.
- Gjaerum HB. The genus Taphrina Fr. in Norway. Nytt Mag Bot. 1964;11:5–26.
- Bacigálová K, Lopandic K, Rodrigues MG, et al. Phenotypic and genotypic identification and phylogenetic characterisation of Taphrina fungi on alder. Mycol Prog. 2003;2(3):179–196.
- Petrydesová J, Bacigálová K, Sulo P. The reassignment of three ‘lost’ Taphrina species (Taphrina bullata, Taphrina insititiae and Taphrina rhizophora) supported by the divergence of nuclear and mitochondrial DNA. Int J Syst Evol Microbiol. 2013;63(Pt 8):3091–3098.
- White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White, TJ, editors. PCR protocols: a guide to methods and applications. San Diego (CA): Academic Press; 1990. p. 315–322.
- Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2(2):113–118.
- Tamura K, Stecher G, Peterson D, et al. MEGA6: molecular evolutionary genetics analysis Version 6.0. Mol Biol Evol. 2013;30(12):2725–2729.
- Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. Version 2.75; 201. Available from: http://mesquiteproject.org
- Ronquist F, Teslenko M, van der Mark P, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542.
- Rambaut A, Drummond A. FigTree v1. 3.1: tree figure drawing tool. Institute of evolutionary biology. Edinburgh: 2009. Available from: http://tree.bio.ed.uk/software/figtree.