Abstract
Penicillium, the most common genus plays an important ecological role in various terrestrial and marine environments. However, only a few species have been reported from rhizosphere soil. As part of a project to excavate Korean indigenous fungi, we investigated rhizosphere soil of six plants in the forest (terrestrial habitat) and sand dunes (coastal habitat) and focused on discovering Penicillium species. A total of 64 strains were isolated and identified as 26 Penicillium species in nine sections based on morphological characteristics and the sequence analysis of β-tubulin and calmodulin. Although this is a small-scale study in a limited rhizosphere soil, eight unrecorded species and four potential new species have been identified. In addition, most Penicillium species from rhizosphere soil were unique to each plant. Penicillium halotolerans, P. scabrosum, P. samsonianum, P. jejuense, and P. janczewskii were commonly isolated from rhizosphere soil. Eight Penicillium species, P. aurantioviolaceum, P. bissettii, P. cairnsense, P. halotolerans, P. kananaskense, P. ortum, P. radiatolobatum, and P. verhagenii were recorded for the first time in Korea. Here, we provide the detailed morphological description of these unrecorded species.
Keywords:
1. Introduction
Fungi play an important role in rhizosphere soil. They affect plant fitness and adaptability in ecosystems through various biological processes [Citation1,Citation2]. The genus Penicillium is one of the most common fungi found in rhizosphere soil [Citation3]. Some Penicillium species produce solubilized phosphorus, siderophore, and phytohormones such as indole acetic acid and gibberellic acid, which are important for plant health [Citation4–6]. Although the importance of Penicillium isolated in rhizosphere soil has been reported, many Penicillium species were only identified at the genus level [Citation7,Citation8]. There are two possible reasons for this; one being that studies conducted previously have focused on simply screening for useful enzymes and bioactive compounds, and two that it is naturally difficult to identify Penicillium species.
The members of the Penicillium genus are easily identified at the genus level based on their microscopic features in culture condition, whereas they are difficult to identify at the species level due to their morphological plasticity under different growth conditions [Citation9]. Recently, the identification of Penicillium species has become much easier since standardized methods including morphology and multigene phylogenetic analysis have been proposed [Citation9]. To date, Penicillium has been reported in 25 sections of 345 species worldwide through the use of standardized methods [Citation9]. In Korea, approximately 102 Penicillium species from terrestrial environments [Citation10,Citation11] and 100 Pencillium species from marine environments [Citation12–17] have been reported. It was confirmed that many Pencillium species from marine environments are different from those found in terrestrial environments. So far, a total of 152 Penicillium species have been reported in Korea. However, there are only a few studies on Penicillium from rhizosphere soil [Citation18,Citation19], and since rhizosphere soil is unique for each plant species, it can be assumed that different Penicillium species may exist for each plant.
As part of the projects organized by the National Institute of Biological Resources (NIBR) and the Ministry of Ocean and Fisheries established the Marine Fungal Resource Bank (MFRB) to excavate Korean indigenous fungi, we investigated rhizosphere soil to discover Penicillium species. The main objective of this study was to isolate Penicillium species from rhizosphere soil of six plants: Rhododendron brachycarpum, Sorbus commixta, and Taxus cuspidate from a forest (terrestrial habitat) and Calystegia soldanella, Lathyrus japonicus, and Orobanche coerulescens from a sand dune (coastal habitat). All Penicillium isolates were identified at the species level using the sequence analysis of the β-tubulin (BenA) and calmodulin (CaM) loci. We identified eight unrecorded species and four new species candidates of Penicillium in Korea.
2. Materials and methods
2.1. Sample collections and isolation
Rhizosphere soil from six plants were collected from six sites in Korea in 2019 (, ). Rhododendron brachycarpum, Sorbus commixta, and Taxus cuspidate were collected from a forest (terrestrial habitat) and Calystegia soldanella, Lathyrus japonicus, and Orobanche coerulescens were collected from a sand dune (coastal habitat). All samples were stored at 4 °C before fungal isolation. For each sample, 5 g of rhizosphere soil was diluted ten-fold with sterile water. Next, 100 μL of each dilution was plated on dichloran rose bengal chloramphenicol agar (Difco, Becton Dickinson, Sparks, MD). All plates were incubated at 25 °C for seven days. Penicillium strains were transferred to a potato dextrose agar (PDA; Difco, Becton Dickinson) plate. Each strain was then stored in 20% glycerol at −80 °C at the Seoul National University Fungus Collection (SFC) ().
Figure 1. The number of Penicillium species from each sampling sites. (A) Map showing the number of (strains/species) at each location. (B) Total, unique, and shared Penicillium species from forest and sand dune. (C) Total, unique, and shared Penicillium species obtained from rhizosphere soil of six different plants.
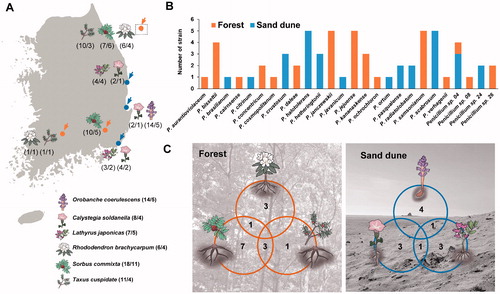
Table 1. Summary and GenBank accession numbers for Penicillium strains isolated from the rhizosphere soil of six different plants. The unrecorded Penicillium species in Korea are represented in bold.
2.2. DNA extraction, amplification, and sequencing
Genomic DNA was extracted from Penicillium using a modified cetyltrimethylammonium bromide extraction protocol [Citation20] with respect to the amount of sample tissue. The primer sets, Bt2a/Bt2b [Citation21] and CF1/CF4 [Citation22] were used to amplify BenA and CaM, respectively. Each PCR was performed in a C1000 thermal cycler (Bio-Rad, Richmond, CA) using previously described methods [Citation14]. The PCR products were purified with a ExpinTM PCR Purification Kit (GeneAll Biotechnology, Seoul, Korea), according to the manufacturer’s instructions. DNA sequencing was performed on an ABI Prism 3700 genetic analyzer (Life Technologies, Gaithersburg, MD) at Macrogen (Seoul, Korea), in both directions using the PCR primers.
2.3. Phylogenetic analysis
The sequences were assembled and proofread using MEGA5 [Citation23] and were deposited in GenBank (accession Nos. are shown in ). Molecular identification was performed in two steps. First, the sectional position of the strains was determined by comparison to the BenA sequences with database containing the sequence of the type strains. Next, each strain was identified to the species level by analyzing the combined dataset of the two loci (BenA and CaM). Talaromyces flavus NRRL 2098 was used as the outgroup [Citation24]. Multiple alignments were performed using the L-INS-i option of MAFFT v7 [Citation25]. Maximum likelihood phylogenetic analyses were performed with RAxML [Citation26] implemented on CIPRES web portal [Citation27], using the GTR + G model with 1000 bootstrap replicates.
2.4. Morphological analysis
The morphology of eight unrecorded species was observed using previously described methods [Citation9] on three different culture media: Czapek yeast autolysate agar (CYA; Difco, Sparks, MD), malt extract agar (MEA; Oxoid, Hampshire, UK), and yeast extract sucrose agar (YES; Difco). The Methuen Handbook of Color was used for the color names and alphanumeric codes for macromorphological characteristics [Citation28]. The microscopic features were observed under a light microscope (Eclipse 80i, Nikon, Tokyo, Japan) using colonies grown on MEA.
3. Results
3.1. Species identification
A total of 64 Penicillium strains were isolated from rhizosphere soil of six plants. They were clustered in 26 groups based on their BenA sequences (Supplemental Figure 1). For identification at the species level, one to six representative strains from each group were used for a phylogenetic analysis using the combined dataset from BenA and CaM. Finally, these were confirmed as 26 species in nine sections including eight unrecorded species and four potential new species (, ).
Figure 2. Maximum likelihood phylogenetic tree of the combined data set of BenA and CaM used to identify strains to the species level in Penicillium from rhizosphere soil. Bootstrap scores of >70 are presented at the nodes. The scale bar indicates the number of nucleotide substitutions per site. “T” indicates the ex-type strains. Strains reported in the current study are represented in bold. The species labeled in red represent previously unrecorded species in Korea. The names in blue are potential new species.
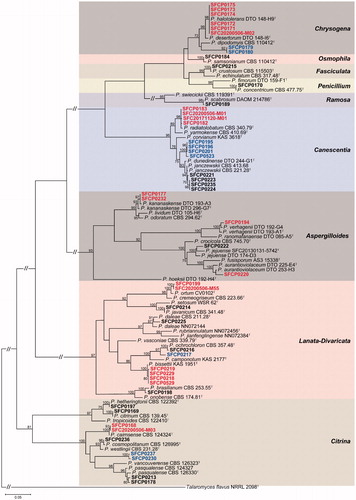
Section Lanata-Divaricata (7 Penicillium spp.) and Citrina (6) showed a relatively higher diversity compared to other sections (). Eight unrecorded species were detected in five sections. These were Aspergilloides (3 spp.), Canescentia (1 sp.), Chrysogena (1 sp.), Citrina (1 sp.), and Lanata-Divaricata (2 spp.). Four potential new species were identified in section Canescentia, Chrysogena, Citrina, and Lanata-Divaricata (). The detailed morphological descriptions for unrecorded species are presented in taxonomic part.
For section Aspergilloides, 10 strains were identified as P. aurantioviolaceum (1 strain), P. jejuense (5), P. kananaskense (3), and P. verhagenii (1). Three species (P. aurantioviolaceum, P. kananaskense, and P. verhagenii) were confirmed as unrecorded species in Korea. For section Canescentia, 11 strains were confirmed as P. janczewskii (5), P. radiatolobatum (2), and Penicillium sp. 04 (4). P. radiatolobatum was an unrecorded species in Korea. Penicillium sp. 04 (SFCP0195, SFCP0196, SFCP0201, and SFCP523) formed a distinct clade from previously reported species. For section Chrysogena, seven strains were determined as P. halotolerans (5) and Penicillium sp. 24 (2). P. halotolerans was an unrecorded species in Korea. Penicillium sp. 24 was a potential new species. For section Citrina, 10 strains were identifed as P. cairnsense (1), P. citrinum (1), P. cosmopolitanum (1), P. hetheringtonii (3), P. pasqualense (2), and Penicillium sp. 26 (2). P. cairnsense was unrecorded species in Korea and Penicillium sp. 26 was a potential new species. Section Lanata-Divaricata contained seven species (11 strains); P. bissettii (4), P. brasilianum (1), P. daleae (2), P. javanicum (1), P. ochrochloron (1), P. ortum (1), and Penicillium sp. 08 (1). Two species (P. bissettii and P. ortum) were new records to Korea. Penicillium sp. 08 (SFCP0217) was a potential new species. P. crustosum (3), P. samsonianum (5), P. concentricum (2), and P. scabrosum (5) included in Section Fasciculata, Osmophila, Penicillium, and Ramosa were identified from rhizosphere soil, respectively (, ).
3.2. Penicillium composition
Penicillium halotolerans, P. scabrosum, P. samsonianum, P. jejuense, and P. janczewskii were the dominant species in rhizosphere soil (). A different number of species were found in the forest (15 spp.) and sand dune (12 spp.). Penicillium sp. 04 was the only species shared across both habitats. Most species were detected in their own sites: forest (14 species) and sand dune (11 species) ().
The different numbers (4–11 Penicillium spp.) of Penicillium species were found depending on the plant surveyed. Penicillium diversity was the highest in Sorbus commixta (11 spp.), followed by Orobanche coerulescens (5) and Lathyrus japonicus (5) (). There were only a few Penicillium species commonly found between the plant hosts on the sand dune, whereas more species were found to overlap in the forest ().
4. Taxonomy
Penicillium aurantioviolaceum Biourge (1923)
Description: Colony diam, 7 d, in mm: CYA 60–65; CYA 15 °C 28–33; CYA 30 °C 28–33; CYA 37 °C no growth; MEA 50–56; YES 60–65 ().
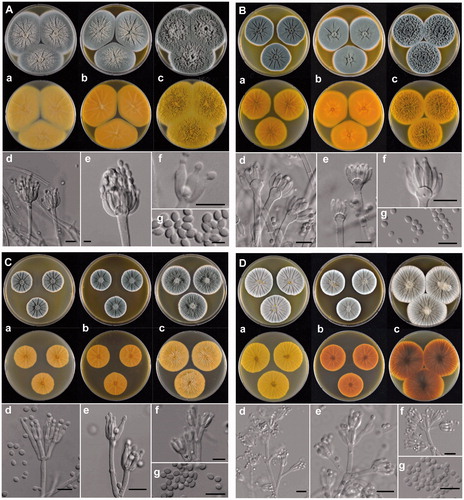
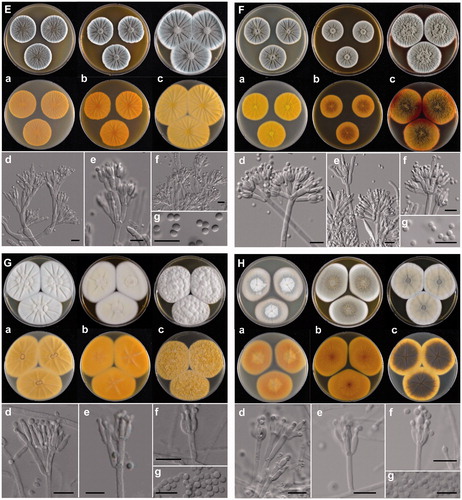
Figure 3. The unrecorded Penicillium species in Korea. (A) P. aurantioviolaceum (SFCP0220), (B) P. bissettii (SFCP0229), (C) P. cairnsense (SFCP0168), (D) P. halotolerants (SFCP0171), (E) P. kananaskense (SFCP0232), (F) P. ortum (SFCP0199), (G) P. radiatolobatum (SFCP0182), (H) P. verhagenii (SFCP0194). (a–c) Colonies grown on Czapek yeast autolysate agar (CYA), malt extract agar (MEA), and yeast extract sucrose agar (YES) from left to right (top = obverse, bottom = reverse). (d–f) Conidiophores; (g) Conidia (scale bar: d–g = 10 μm).
Colony characters: CYA, 25 °C, 7d: Colonies low, radially sulcate; margins low to moderately deep, wide, entire; mycelia white; texture velvety; sporulation dense; conidia dull green (25D3); exudates clear; soluble pigments absent; reverse color pastel yellow (3A4). MEA, 25 °C, 7d: Colonies low to moderately deep, radially sulcate; margins low, wide, entire; mycelia white; texture velvety; sporulation dense; conidia dull green (25D3); exudates clear; soluble pigments absent; reverse color deep yellow (4A8). YES, 25 °C, 7d: Colonies deep, randomly furrowed; margins low, narrow, entire; mycelia white; texture velvety, slightly fasciculate at center; sporulation dense; conidia dull green (25D3); exudates absent; soluble pigments absent; reverse color grayish yellow with light yellow (3A5) at margin.
Conidiophores monoverticillate, rough walls, 2.4–4.0 μm wide, phialides ampulliform, 8.0–11.0 × 2.5–3.0 μm. Conidia smooth walls, ellipsoidal, 3.0–3.7 × 2.0–2.8 μm; Sclerotia white when young becoming orange (6B7) at age; Asci and ascospores not observed.
Strain examined: SFCP0220 and SFC20190612-M12
Note: Penicillium aurantioviolaceum is phylogenetically similar to P. fusisporum. The former species can be distinguished from the latter by the shape of the conidia and growth rate on CYA at 25 °C. P. aurantioviolaceum is characterized by ellipsoidal spores, whereas P. fusisporum has fusiform to oblong conidia. P. aurantioviolaceum grows faster than P. fusisporum on CYA at 25 °C (60–65 vs. 50–53) [Citation29].
Penicillium bissettii Visagie & Seifert (2016)
Description: Colony diam, 7 d, in mm: CYA 45–55; CYA 15 °C 25–30; CYA 30 °C 35–40; CYA 37 °C no growth; MEA 50–60; YES 45–55 ().
Colony characters: CYA, 25 °C, 7d: Colonies low to moderately deep, radially sulcate; margins low, wide, entire; mycelia white; texture floccose; sporulation absent to sparse; exudates clear; soluble pigments absent; reverse color grayish yellow (3B5) with pale yellow (3A3) at margin. MEA, 25 °C, 7d: Colonies low, radially sulcate; margins low, wide, entire; mycelia white; texture floccose; sporulation absent to sparse; conidia grayish green (25B2); exudates clear; soluble pigments absent; reverse color deep yellow (4A8). YES, 25 °C, 7d: Colonies low to moderately deep, randomly furrowed; margins low, narrow, entire; mycelia white; texture floccose; sporulation dense; conidia grayish green (25B2); exudates absent; soluble pigments absent; reverse color grayish yellow (4B4).
Conidiophores biverticillate; stipes rough walls, 2.0–3.0 μm wide, phialides ampulliform, 7.0–11.0 × 2.5–3.5 μm. Conidia rough walls, globose to subglobose, 2.5–3.5 μm; Sclerotia absent; Asci and ascospores not observed.
Strain examined: SFCP0218, SFCP0219, SFCP0229, and SFCP0529
Note: Penicillium bissettii is similar to P. vasconiae and P. annulatum. This species is characterized by the roughened conidiophores and no growth at 37 °C, whereas P. desertorum has smooth walled conidiophores. Penicillium annulatum can be distinguished from P. bissettii by good growth on CYA at 37 °C [Citation30]. Penicillium bissettii in Korea grows faster than type strain of Penicillium bissettii on CYA at 25 °C [Citation30]. The type strain of Penicillium bissettii showed pinkish red mycelia on MEA, whereas the Korean strains have white mycelia.
Penicillium cairnsense Houbraken, Frisvad & Samson (2011)
Description: Colony diam, 7 d, in mm: CYA 36–38; CYA 15 °C 17–19; CYA 30 °C 9–10; CYA 37 °C no growth; MEA 30–33; YES 42–47 ().
Colony characters: CYA, 25 °C, 7d: Colonies low to moderately deep, radially sulcate; margins low, narrow, entire; mycelia white; texture velvety; sporulation dense; conidia dull green (27E3); exudates clear; soluble pigments absent; reverse color light yellow (4A4). MEA, 25 °C, 7d: Colonies low to moderately deep, radially sulcate; margins low, narrow, entire; mycelia white; texture velvety; sporulation dense; conidia dull green (27E3); exudates absent; soluble pigments absent; reverse color light brown (7D5) at center, orange white (5A2) elsewhere. YES, 25 °C, 7d: Colonies low to moderately deep, radially sulcate, randomly furrowed as well, sunken in at center; margins low, narrow, entire; mycelia white; texture velvety; sporulation dense; conidia dull green (27E3); exudates absent; soluble pigments brownish red (9C6); reverse color brownish gray (11E2) with grayish red (8B63) area present.
Conidiophores mostly biverticillate; stipes smooth walls, 2.7–3.7 μm wide, phialides ampulliform, 7.0–10 × 2.5–3.5 μm. Conidia smooth walls, subglobose to broadly ellipsoidal, 2.5–3.5 × 2.5–3.0 μm; Sclerotia absent; Asci and ascospores not observed.
Strain examined: SFCP0168 and SFC20200506-M03
Note: Penicillium cairnsense is morphologically similar to P. quebecense. This species (9–10 mm in length) can be distinguished from P. quebecense (16–20 mm) by slower growth on CYA at 30 °C [Citation31].
Penicillium halotolerans Frisvad, Houbraken & Samson (2012)
Description: Colony diam, 7 d, in mm: CYA 30–35; CYA 15 °C 20–25; CYA 30 °C 30–35; CYA 37 °C 10–13; MEA 34–36; YES 50–54 ().
Colony characters: CYA, 25 °C, 7d: Colonies low to moderately deep, radially sulcate; margins low, narrow, entire; mycelia white; texture velvety; sporulation dense; conidia grayish turquoise (24D3); exudates clear to pale yellow (3A3); soluble pigments absent; reverse color yellowish orange (4B7). MEA, 25 °C, 7d: Colonies low to moderately deep, radially sulcate; margins low, narrow, entire; mycelia white; texture velvety, slightly floccose in center; sporulation moderate; conidia grayish turquoise (24D3); exudates clear to pale yellow (3A3) at center; soluble pigments absent; reverse color brownish orange (5C6). YES, 25 °C, 7d: Colonies low to moderately deep, radially sulcate; margins low, narrow, entire; mycelia white; texture floccose; sporulation moderate; conidia grayish turquoise (24D3), pale turquoise (24A3) at margin; exudates absent; soluble absent; reverse color light yellow (4A4).
Conidiophores terverticillate; stipes smooth walls, 3.0–4.0 μm wide, phialides ampulliform, 6.0–11.0 × 2.5–3.5 μm. Conidia smooth walls, globose, 2.5–3.5 μm; Sclerotia absent; Asci and ascospores not observed.
Strain examined: SFCP0171, SFCP0174, and SFC20200506-M02
Note: Penicillium cairnsense is phylogenetically similar to P. desertorum. This species is characterized by radially sulcate colonies on YES, whereas P. desertorum has randomly furrowed colonies on YES. Penicillium halotolerans in Korea grows faster than the type strain of P. halotolerans on CYA at 30 and 37 °C. (30–35 vs. 20–25 at 30 °C and 10–13 vs. 0–2 at 37 °C) [Citation32].
Penicillium kananaskense Seifert, Frisvad & McLean (1994)
Description: Colony diam, 7 d, in mm: CYA 38–42; CYA 15 °C 24–26; CYA 30 °C 14–16; CYA 37 °C no growth; MEA 42–47; YES 45–48 ().
Colony characters: CYA, 25 °C, 7d: Colonies low to moderately deep, radially sulcate; margins low, narrow, entire; mycelia white; texture velvety; sporulation dense; conidia dull green (25E3); exudates absent; soluble pigments absent; reverse color golden yellow (5B7), deep yellow (4A8) at margin. MEA, 25 °C, 7d: Colonies low, radially sulcate; margins low, narrow, entire; mycelia white; texture velvety; sporulation dense; conidia dull green (25D3); exudates absent; soluble pigments absent; reverse color deep orange (5A8). YES, 25 °C, 7d: Colonies moderately deep, randomly furrowed; margins low, narrow, entire; mycelia white; texture velvety; sporulation dense; conidia dull green (25D3); exudates absent; soluble pigments absent; reverse color dark yellow (4C8) with orange yellow (4A7) at margin.
Conidiophores monoverticillate; stipes smooth walls or rough walls, 3.0–4.5 μm wide, phialides ampulliform, 8.0–13.0 × 3.0–4.0 μm. Conidia rough walls, broadly ellipsoidal to ellipsoidal, 3.0–3.5 × 2.5–3.0 μm; Sclerotia absent; Asci and ascospores not observed.
Strain examined: SFCP0176, SFCP0177, and SFCP0232
Note: Penicillium kananaskense is phylogenetically similar to P. lividum and P. odoratum. This species is characterized by velvety colonies on MEA and slower growth on CYA at 30 °C [Citation33].
Penicillium ortum Visagie & K. Jacobs (2015)
Description: Colony diam, 7 d, in mm: CYA 39–46; CYA 15 °C 20–23; CYA 30 °C 38–44; CYA 37 °C 24–32; MEA 50–55; YES 45–50 ().
Colony characters: CYA, 25 °C, 7d: Colonies low to moderately deep, radially sulcate; margins low, narrow, entire; mycelia white at center, orange white (6A2) at margin; texture floccose; sporulation absent to sparse; conidia greenish gray (26E2); exudates clear; soluble pigments absent; reverse color pale orange (5A3) at center, fading into brownish orange to pale yellow (4A3). MEA, 25 °C, 7d: Colonies low, plane; margins low, narrow, entire; mycelia white to light yellow; texture velvety to floccose; sporulation moderate; conidia greenish gray (26E2); exudates absent; soluble pigments absent; reverse color brown (6D7) at center, brownish orange (5C6) elsewhere. YES, 25 °C, 7d: Colonies moderately deep, radially sulcate; margins low, narrow, entire; mycelia white at margin, light yellow elsewhere; texture floccose; sporulation sparse; conidia greenish gray (26E2); exudates absent; soluble pigments absent; reverse color pale yellow at margin, grayish brown (6E3) elsewhere.
Conidiophores mostly biverticillate, sometimes monoverticillate; stipes smooth walls, 2.0–3.2 μm wide, phialides ampulliform, 6.5–9.5 × 2.2–3.2 μm. Conidia smooth walls, globose to subglobose, 2.5–3.3 × 2.4–3.1 μm; Sclerotia absent; Asci and ascospores not observed.
Strain examined: SFCP0199 and SFC20200506-M55
Note: Penicillium ortum is phylogenetically similar to P. cremeogriseum. The former species can be distinguished from the latter by relatively fast growth on CYA at 30 °C (38–44 mm) [Citation34].
Penicillium radiatolobatum Lörinczi (1972)
Description: Colony diam, 7 d, in mm: CYA 35–40; CYA 15 °C 18–21; CYA 30 °C 28–33; CYA 37 °C 10–13; MEA 30–35; YES 45–51 ().
Colony characters: CYA, 25 °C, 7d: Colonies low to moderately deep, radially sulcate; margins low, narrow, entire; mycelia white; texture velvety, slightly floccose at central; sporulation moderate; conidia greenish gray (25D2); exudates pale yellow (2A3) droplets at central; soluble pigments absent; reverse color dull yellow (3B3) to olive brown (4B3) at margin. MEA, 25 °C, 7d: Colonies low, radially sulcate; margins low, narrow, entire; mycelia white; texture floccose; sporulation moderate; conidia dull green (28E3); exudates hyaline to orange white (5A2) droplets in central areas; soluble pigments absent; reverse color light brown (6D5). YES, 25 °C, 7d: Colonies low, radially sulcate; margins low, narrow, entire; mycelia white; texture velvety to floccose; sporulation moderate; conidia greenish gray (25D2); exudates absent; soluble pigments absent; reverse color brown (7E7).
Conidiophores biverticillate; stipes smooth walls, 2.8–4.0 μm wide, phialides ampulliform, 5.5–9.0 × 2.5–3.0 μm. Conidia smooth walls, globose to subglobose, 2.8–3.2 × 2.8–3.1 μm; Sclerotia absent; Asci and ascospores not observed.
Strain examined: SFCP0182, SFCP0183, SFC20171120-M01, and SFC20200506-M01
Note: Penicillium radiatolobatum is phylogenetically similar to P. canescens. This species can be distinguished from P. canescens by faster growth on CYA and MEA at 25 °C [Citation30,Citation35].
Penicillium verhagenii Houbraken (2014)
Description: Colony diam, 7 d, in mm: CYA 27–30; CYA 15 °C 24–26; CYA 30 °C no growth; CYA 37 °C no growth; MEA 28–31; YES 34–40 ().
Colony characters: CYA, 25 °C, 7d: Colonies low to moderately deep, radially sulcate; margins low, narrow, entire; mycelia white; texture velvety to floccose; sporulation moderate; conidia grayish green (27C3); exudates absent; soluble pigments absent; reverse color pale yellow (4A3) to grayish yellow (4B5). MEA, 25 °C, 7d: Colonies low to moderately deep, radially sulcate; margins low, narrow, entire; mycelia white; texture velvety to floccose; sporulation moderate; conidia dull green (25E4); exudates absent; soluble pigments absent; reverse color pale brownish orange (5C4). YES, 25 °C, 7d: Colonies low to moderately deep, radially sulcate, random furrows also present; margins low, narrow, entire; mycelia white; texture velvety to floccose; sporulation moderate; conidia dull green (27E3) at the center of colony, but greenish gray (27B2) at margin; exudates absent; soluble pigments absent; reverse color grayish yellow (4B3).
Conidiophores biverticillate; stipes finely roughoth walls, 2.8–3.8 μm wide, phialides ampulliform, 8.0–11.0 × 2.8–3.5 μm. Conidia roughened walls, broadly ellipsoidal to ellipsoidal, 3.0–4.0 × 2.5–3.5 μm; Sclerotia absent; Asci and ascospores not observed.
Strain examined: SFCP0194
Note: Penicillium verhagenii is phylogenetically similar to P. ranomafanaense. P. verhagenii is characterized by yellow reverse colors on CYA and YES, whereas P. ranomafanaense has orange or reddish [Citation33].
5. Discussion
The rhizosphere is known as the most dynamic environment that provides a close association between plant root and fungi [Citation1]. We isolated 64 Penicillium strains from rhizosphere soil of six plants. They were identified accurately by sequence-based identification as 26 species in nine sections. Four species could not be identified due to an unclear phylogenetic relationship. They were designated as Penicillium sp. We might be able to report them as new species in the future after a more detailed morphological comparison with phylogenetically similar species.
Eight species were records for the first time in Korea. These were P. aurantioviolaceum, P. bissettii, P. cairnsense, P. halotolerans, P. kananaskense, P. ortum, P. radiatolobatum, and P. verhagenii. The morphological characteristics of the unrecorded species were consistent with those of the respective type species, except for P. bissettii and P. halotolerans. The strains isolated from Korea of these two species grow faster compared to the type strains. Some fungi exhibit different growth rates or metabolism as they adapt to different environments [Citation36,Citation37]. P. bissettii and P. halotolerans were isolated from sand dune in Korea, whereas their type cultures were obtained from forest soil and salt marsh, respectively [Citation30,Citation32]. Although this is a small-scale study in a limited habitat, many unrecorded species and potential new species have been found. By analyzing a larger variety of hosts, it would be possible to discover more species.
Fungal diversity and composition were significantly correlated with habitat and plant communities [Citation38,Citation39]. Fungal diversity was significantly higher in terrestrial habitats than freshwater and mangrove habitats [Citation38]. Similarly, the diversity of Penicillium species in rhizosphere soil was relatively higher in terrestrial habitats than coastal habitats. Most Penicillium species from rhizosphere soil were unique to each plant. The composition of the Penicillium species did not only differ by habitats but also by the plant host species. The Penicillium composition, in particular, is relatively much affected by plants in the marine environment. Despite the two host plants, Calystegia soldanella and Lathyrus japonicus having very similar environments, such as poor nutrient and abiotic stresses, fungal diversity varied depending on plants [Citation40]. Recently, various Penicillium species have been reported by NGS-based studies in rhizosphere soil [Citation41,Citation42]. Although the role of these Penicillium in rhizosphere soil is unclear, they might have important effects on plants. Further studies are required to investigate the function of Penicillium in rhizosphere soil.
Supplemental Material
Download GIF Image (646.5 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Coats VC, Rumpho ME. The rhizosphere microbiota of plant invaders: an overview of recent advances in the microbiomics of invasive plants. Front Microbiol. 2014;5:368
- Ehrmann J, Ritz K. Plant: soil interactions in temperate multi-cropping production systems. Plant Soil. 2014;376(1-2):1–29.
- Workneh F, Van Bruggen AHC. Microbial density, composition, and diversity in organically and conventionally managed rhizosphere soil in relation to suppression of corky root of tomatoes. Appl Soil Ecol. 1994;1(3):219–230.
- Altaf MM, Imran M, Abulreesh HH, et al. Diversity and applications of Penicillium spp. in plant-growth promotion. In: Gupta, V.K., & Rodriguez-Couto, S. (eds.), New and Future Developments in Microbial Biotechnology and Bioengineering: Penicillum System Properties and Applications. Elsevier. Amsterdam, Netherlands; 2017. p. 261–276.
- Berg G, Zachow C, Lottmann J, et al. Impact of plant species and site on rhizosphere-associated fungi antagonistic to Verticillium dahliae Kleb. Appl Environ Microbiol. 2005;71(8):4203–4213.
- Elias F, Woyessa D, Muleta D. Phosphate solubilization potential of rhizosphere fungi isolated from plants in Jimma Zone, Southwest Ethiopia. Int J Microbiol. 2016;2016:5472601
- An YN, Zhang X, Zhang TY, et al. Penicimenolides AF, resorcylic acid lactones from Penicillium sp., isolated from the rhizosphere soil of panax notoginseng. Sci Rep. 2016;6:27396.
- Rai R, Kaur B, Singh S, et al. Evaluation of secretome of highly efficient lignocellulolytic Penicillium sp. Dal 5 isolated from rhizosphere of conifers. Bioresour Technol. 2016;216:958–967.
- Visagie CM, Houbraken J, Frisvad JC, et al. Identification and nomenclature of the genus Penicillium. Stud Mycol. 2014a;78:343–371.
- Kim HJ, Kim JS, Cheon KH, et al. Species list of Aspergillus, Penicillium and Talaromyces in Korea, based on ‘One Fungus One Name’ system. Kor J Mycol. 2016;44:207–219.
- National List of Species of Korea. 2019. National Institute of Biological Resources, online at http://kbr.go.kr., accessed on 13 July 2020.
- Park MS, Fong JJ, Oh SY, et al. Marine-derived Penicillium in Korea: diversity, enzyme activity, and antifungal properties. Antonie Van Leeuwenhoek. 2014;106(2):331–345.
- Park MS, Eom JE, Fong JJ, et al. New record and enzyme activity of four species in Penicillium section Citrina from marine environments in Korea. J Microbiol. 2015;53(4):219–225.
- Park MS, Lee S, Oh SY, et al. Diversity and enzyme activity of Penicillium species associated with macroalgae in Jeju Island. J Microbiol. 2016;54(10):646–654.
- Park MS, Lee S, Lim YW. A New record of four Penicillium species isolated from Agarum clathratum in Korea. J Microbiol. 2017;55(4):237–246.
- Park MS, Oh SY, Lee S, et al. Fungal diversity and enzyme activity associated with sailfin sandfish egg masses in Korea. Fungal Ecol. 2018;34:1–9.
- Park MS, Oh SY, Fong JJ, et al. The diversity and ecological roles of Penicillium in intertidal zones. Sci Rep. 2019;9(1):1–11.
- Park MS, Yu SH. Plant growth promoting fungi isolated from rhizosphere of zoysiagrass in Korea. Kor J Mycol. 2005;33:30–34.
- Babu AG, Kim SW, Yadav DR, et al. Penicillium menonorum: a novel fungus to promote growth and nutrient management in cucumber plants. Mycobiology. 2015;43(1):49–56.
- Rogers SO, Bendich AJ. Extraction of total cellular DNA from plants, algae and fungi. In: S. Gelvin and R. Schilperoort (eds.), Plant molecular biology manual., Kluwer Academic, Dordrecht; 1994.
- Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 1995;61(4):1323–1330.
- Peterson SW, Vega FE, Posada F, et al. Penicillium coffeae: a new endophytic species isolated from a coffee plant and its phylogenetic relationship to P. fellutanum, P. thiersii and P. brocae based on parsimony analysis of multilocus DNA sequences. Mycologia. 2005;97(3):659–666.
- Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739.
- Visagie CM, Hirooka Y, Tanney JB, et al. Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world. Stud Mycol. 2014b;78:63–139.
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780.
- Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690.
- Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES science gateway for inference of large phylogenetic trees. SC10 workshop on gateway computing environments (GCE10), New Orleans, LA; 2010. p.1–8.
- Kornerup A, Wanscher JH. Methuen handbook of colour. 3rd ed. Methuen. London; 1978.
- Wang B, Yu Y, Wang L. Penicillium fusisporum and P. zhuangii, two new monoverticillate species with apical-swelling stipes of section Aspergilloides isolated from plant leaves in China. PloS One. 2014;9(7):e101454.
- Visagie CM, Renaud JB, Burgess KMN, et al. Fifteen new species of Penicillium. Persoonia. 2016;36:247–280.
- Houbraken J, Frisvad JC, Samson RA. Taxonomy of Penicillium section Citrina. Stud Mycol. 2011;70(1):53–138.
- Houbraken J, Frisvad JC, Seifert KA, et al. New penicillin-producing Penicillium species and an overview of section Chrysogena. Pers - Int Mycol J. 2012;29(1):78–100.
- Houbraken J, Visagie CM, Meijer M, et al. A taxonomic and phylogenetic revision of Penicillium section Aspergilloides. Stud Mycol. 2014;78:373–451.
- Visagie CM, Houbraken J, Seifert KA, et al. Four new Penicillium species isolated from the fynbos biome in South Africa, including a multigene phylogeny of section Lanata-Divaricata. Mycol Prog. 2015;14(10):96.
- Pitt JI. The genus Penicillium and its teleomorphic states Eupenicillium and Talaromyces. Academic Press, London, UK; 1979.
- Bidochka M, Menzies F, Kamp A. Genetic groups of the insect-pathogenic fungus Beauveria bassiana are associated with habitat and thermal growth preferences. Arch Microbiol. 2002;178(6):531–537.
- Baakza A, Vala AK, Dave BP, et al. A comparative study of siderophore production by fungi from marine and terrestrial habitats. J Exp Mar Biol Ecol. 2004;311(1):1–9.
- Vijaykrishna D, Jeewon R, Hyde KD. Molecular taxonomy, origins and evolution of freshwater ascomycetes. Fungal Divers. 2006;23:351–390.
- Gao C, Shi NN, Chen L, et al. Relationships between soil fungal and woody plant assemblages differ between ridge and valley habitats in a subtropical mountain forest. New Phytol. 2017;213(4):1874–1885.
- Quilliam RS, Jones DL. Evidence for host-specificity of culturable fungal root endophytes from the carnivorous plant Pinguicula vulgaris (Common Butterwort). Mycol Progress. 2012;11(2):583–585.
- Qiao Q, Zhang J, Ma C, et al. Characterization and variation of the rhizosphere fungal community structure of cultivated tetraploid cotton. PloS One. 2019;14(10):e0207903
- Floc’h JB, Hamel C, Harker KN, et al. Fungal communities of the canola rhizosphere: keystone species and substantial between-year variation of the rhizosphere microbiome. Microbial Ecol. 2020. DOI:10.1007/s00248-019-01475-8