Abstract
Acervus (Pyronemataceae, Pezizales) is a saprobic genus in Pezizomycetes, characterized by colored apothecia, subcylindrical to cylindrical asci and guttulate ascospores. We collected four Acervus samples from China and Thailand. Descriptions and illustrations are introduced for all fresh samples. One new record of A. globulosus from Thailand, one new species, A. rufus, two known species, A. epispartius and A. stipitatus from China are reported. Phylogenetic analysis based on five genes, the large subunit rRNA (LSU), the translation elongation factor-1 alpha (tef1-α), the second largest subunit of RNA polymerase II (rpb2), the largest subunit of RNA polymerase II (rpb1), and the small subunit rRNA (SSU), revealed the distinct position of the new species. The new species is set apart by its red apothecia. A key to Acervus species is also given.
1. Introduction
Acervus Kanouse includes nine species and is characterized by sessile or stipitate, discoid to cup-shaped or pulvinate, vivid colored apothecia, inamyloid asci, and guttulate ascospores [Citation1–3]. The type species of the genus is Acervus aurantiacus Kanouse, which was collected from soil under an elm tree in the USA [Citation4]. When Kanouse [Citation4] examined this species, he did not observe operculate asci and placed the genus within Dermateaceae. Seaver [Citation5] moved Acervus to the tribe Ascotremelleae in Helotiaceae owing to its rubbery sclerotiform base and inoperculate asci. Korf [Citation6] reexamined type material and placed Acervus in Sarcosomataceae. Korf [Citation7] placed Acervus into Pyronemataceae on account of its thin-walled asci, while Denison [Citation8] treated Acervus in Sarcosyphaceae. Pfister [Citation9] included Phaedropezia Le Gal in the genus and proposed two combinations: Acervus epispartius (Berk. & Broome) Pfister (= Phaedropezia epispartia (Berk. & Broome) Le Gal) and Acervus flavidus (Berk. & M.A. Curtis) Pfister (= Phaedropezia flavida (Berk. & M.A. Curtis) Le Gal). Upon examination of holotype of A. aurantiacus and P. epispartia, Pfister [Citation9] concluded that the two species have no notable differences. Due to P. epispartia taking precedence over A. aurantiacus, the names of the two species were combined as A. epispartius, which is the current name for the type species in Acervus [Citation9]. Moravec [Citation10] provided descriptions of his collection of A. epispartius and of a new species, Acervus lusakianus J. Moravec. Moravec [Citation10] observed that the A. epispartius he collected was identical to P. epispartia, but congeneric with Acervus. Based on previous discussions on Acervus and Phaedropezia, he adopted Pfister’s recommendations regarding the emendation of Acervus [Citation10].
For a long time, the establishment of new species depended only on morphology. For example, Zhuang and Wang collected Acervus xishuangbannicus W.Y. Zhuang and Zheng Wang in rotten wood from tropical China and provided morphological description [Citation11]. Nonetheless, the classification of Acervus was varied because the views of various researchers differed [Citation4–8,Citation12]. This was illustrated when an examination of the same material resulted in different classification schemes due to alternative points of view [Citation6,Citation9,Citation12]. Initial phylogenetic analysis placed Acervus within Pyronemataceae [Citation13] and Acervus species formed a highly-supported monophyletic group. Zhuang et al. [Citation1] introduced A. beijingensis W.Y. Zhuang and A. changchunensis W.Y. Zhuang from China, using evidence from morphology and phylogeny. Acervus heilongjiangensis F. Ren & W.Y. Zhuang was collected from soil from a temperate area in China [Citation2]. Shortly afterwards, Acervus globulosus Ekanayaka, Q. Zhao & K.D. Hyde and A. stipitatus Ekanayaka, Q. Zhao & K.D. Hyde were described and a detailed comparative table of morphological features of Acervus species was provided [Citation3]. Subsequently, extensive phylogenetic analysis placed Acervus in Otideaceae within the Pezizomycetes, along with eight additional genera [Citation3,Citation14,Citation15]. In the updated outline of fungi, Otideaceae is synonymized to Pyronemataceae [Citation16].
Herein, we introduce four new collections from China and Thailand. To obtain an insight into the phylogenetic position of these morphologically similar species, sequences of the large subunit rRNA (LSU), translation elongation factor 1-alpha (tef1-α), RNA polymerase II second largest subunit (rpb2), RNA polymerase II largest subunit (rpb1) and small subunit rRNA (SSU) were analyzed jointly with sequences of closely related taxa within Acervus. We introduce a new species of Acervus, provide a detailed account of morphological characteristics for all collections, as well as a key to species of the genus.
2. Materials and methods
2.1. Sample collection, morphological studies, and deposition
Four fresh samples were collected from soil and humus. Three samples were collected from Yunnan Province, southwest China, and the rest one was collected from Chiang Mai Province, Thailand. These samples were brought to the laboratory in plastic bags containing allochroic silica gel. In addition, we loaned an Herbarium specimen (MFLU 16-0607) from the Herbarium of Mae Fah Luang University (MFLU). Samples and herbarium were observed using a Motic SMZ-168 stereoscope and photographed by Nikon Eclipse 80i compound microscope with a Cannon EOS 600 D camera. Cotton Blue, Melzer’s, Gongo Red, and 5% KOH, was used to stain and observe morphological characteristics. Measurements were made using the Tarosoft® Image Frame Work program v. 0.9.7. New herbarium specimens were deposited at the Herbarium of Mae Fah Luang University (MFLU) and the Herbarium of Cryptogams Kunming Institute of Botany Academia Sinica (HKAS). Facesoffungi and Index Fungorum numbers were obtained as in Jayasiri et al. [Citation17] and Index Fungorum (2020).
2.2. DNA extraction, PCR amplification, and sequencing
Genomic DNA of fresh samples was extracted directly from ascomata using a TreliefTM Plant Genomic DNA Kit. The large subunit rRNA (LSU), translation elongation factor 1-alpha (tef1-α), RNA polymerase II second largest subunit (rpb2), RNA polymerase II largest subunit (rpb1), and small subunit rRNA (SSU) regions were amplified by polymerase chain reaction (PCR) using the primers listed in . Total reaction volume was 25 μl containing 9.5 μl sterile deionized water, 12.5 μl of 2 × Power Taq PCR MasterMix, 1 μl of each primer (10 μM stock) and 1 μl of DNA template. Amplifications were carried out using an Applied Biosystems 2720 thermocycler (Foster City, CA, USA). Conditions of PCR for each gene are shown in . PCR products were visualized using 1% agarose gel electrophoresis and distinct bands viewed in Gel documentation system. The PCR products were purified using a TreliefTM DNA Gel Extraction Kit and sequenced by Tsingke Company, Beijing, P.R. China.
Table 1. Primers used for amplification.
Table 2. PCR conditions used for amplification.
2.3. Phylogenetic analysis
Relevant sequences from GenBank are listed in [Citation1–3,Citation13]. Sequences were assembled using BioEdit v.7.0.5.3 [Citation23] and aligned using MAFFT v.7.110 online program (http://mafft.cbrc.jp/alignment/server/) [Citation24]. Dataset was trimmed by TrimAl v.1.3 using the gappyout option (http://phylemon.bioinfo.cipf.es/utilities.html) [Citation25].
Table 3. Sequences used in this study.
A maximum likelihood (ML) phylogenetic analysis was carried out by RAxMLGUI v.1.3 [Citation26] under the GTR + GAMMA nucleotide substitution model with 1000 rapid bootstrap replicates.
Maximum-parsimony analysis was performed using PAUP v.4.0b10 [Citation26]. Gaps were treated as missing data with the heuristic search option with 1000 random sequence additions and tree bisection reconnection (TBR) branch-swapping. Maxtrees were unlimited, branches of zero length were collapsed and all parsimonious trees were saved. Clade stability was assessed using a bootstrap (BT) analysis with 1000 replicates, each with 10 replicates of random stepwise addition of taxa.
Bayesian inference (BI) analysis was performed using MrBayes v.3.2.6 [Citation27]. Model of evolution was estimated by using MrModeltest v.2.3 [Citation28] as performed by MrMTgui [Citation29] based on the Akaike information criterion [Citation30]. Posterior probabilities (PP) [Citation31] were calculated by Markov Chain Monte Carlo Sampling (MCMC) [Citation32]. Six simultaneous Markov Chains were run for 50,000,000 generations and tree sampling occurred every 100th generations [Citation33]. 25% of these trees were discarded as burn-in and analysis was stopped when the standard deviation of split frequency reached 0.01.
Alignment and trees were submitted to TreeBASE (submission ID: 25938). New sequences have been submitted to GenBank and accession numbers have been obtained ().
3. Results
3.1. Phylogenetic analyses
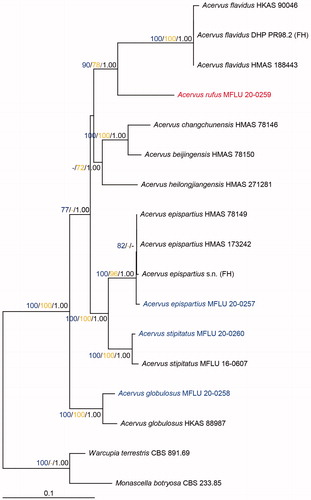
The alignment contained 17 taxa spanning the current molecular diversity of Acervus along with Monascella botryosa Guarro & Arx (CBS 233.85) and Warcupia terrestris Paden & J.V. Cameron (CBS 891.69) as outgroup taxa. The combined matrix of LSU, tef1-α, rpb2, rpb1, and SSU included 5257 sites (LSU: 1 − 906 bp, tef1-α: 907 − 1872 bp, rpb2: 1873 − 2763 bp, rpb1: 2764 − 3550 bp, SSU: 3551 − 5257 bp). Of these, 4212 characters were constant and 705 were parsimony informative. The phylogenetic tree can be seen in . Likelihood value of the best scoring RaxML tree was −16128.204161. The most parsimonious trees (MPTs) showed tree length of 1908 steps [CI = 0.680, RI = 0.632, RC = 0.430, HI = 0.320]. Acervus is monophyletic and this relationship has maximum support with all three methods of analysis (100ML/100MP/1.00BI). Acervus epispartius (MFLU 20-0257) grouped with the rest of the isolates of this species with strong support (100ML/96MP/1.00BI). Acervus stipitatus (MFLU 20-0260) and A. globulosus (MFLU 20-0258) grouped with isolates of the same species also with maximum support (100ML/100MP/1.00BI). Acervus rufus (MFLU 20-0258) grouped as sister to A. flavidus strains. The clade support was 90ML/78MP/1.00BI. Acervus rufus (MFLU 20-0258) and A. flavidus (HKAS 188443), differ by 61 base pairs including 9 gaps in the LSU region (888 bp), 47 base pairs including 3 gaps in the tef1-α region (432 bp), 150 base pairs without gaps in the rpb2 region (897 bp) and 68 base pairs including 4 gaps in the rpb1 region (594 bp).
Figure 1. Phylogenetic tree of combined LSU, tef1-α, rpb2, rpb1, and SSU sequence data inferred from 17 taxa and 5257 sites under the GTR (general time reversible) + GAMMA model of nucleotide substitution. Numerical values at the nodes indicate maximum likelihood bootstrap support (blue), maximum parsimony bootstrap support (yellow), and posterior probabilities (black). Only values greater than 70% (maximum likelihood and maximum parsimony) and 0.95 (Bayesian analysis) are indicated. Name in red indicates new species. Names in blue indicate new collections. Tree is artificially rooted to Monascella botryosa (CBS 233.85) and Warcupia terrestris (CBS 891.69) taxa.
3.2. Taxonomy
Acervus epispartius (Berk. & Broome) Pfister, Occ. Pap. Farlow Herb. Crypt. Bot. 8: 3 (1975) ()
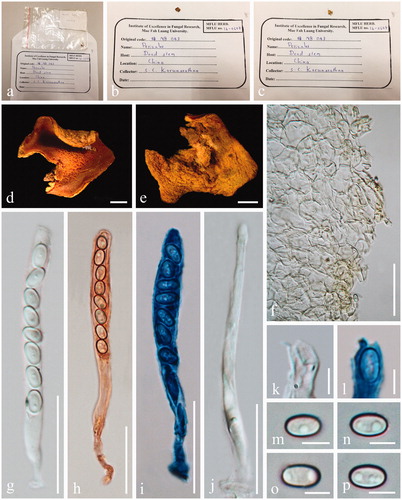
Figure 2. Acervus epispartius (HKAS 107301). (a–d) Typical mature specimens. (e) Receptacle surface of pileus in 5% KOH. (f) Asci and paraphyses in 5% KOH. (g, h) Paraphyses in Congo red. (i, j) Asci. (j Asci in Congo red.). (k, l) Apexes of asci. (k Apex in Congo red. l Apex in Cotton blue.). m-p Ascospores. Scale bars: e–f = 50 μm. g–h = 20 μm. i–j = 30 μm. k–p = 5 μm.

Index Fungorum No: IF 308081
Description: Saprobic on soil and humus. Sexual morph: Apothecia 20–50 mm broad, scattered to gregarious, arising from a yellowish mycelia pad, forming a short stalked, up to 3 mm long, cupulate to pulvinate, hymenium yellow, receptacle surface yellow. Medullary excipulum 440 − 650 µm broad, of textura intricata, yellowish, composed of 7–11 µm broad hyphae. Ectal excipulum 90 − 195 µm broad, of textura angularis to textura globulosa, mix with textura epidermoidea, yellowish, composed of 20 − 28 × 16 − 24 µm broad cells, slightly pubescent. Paraphyses 4 − 5 µm diam, yellowish, branched or unbranched, septate, filiform. Asci 79 − 98 × 5 − 6 µm, 8-spored, operculate, subcylindrical, J-. Ascospores [20/1/1, in 5% KOH] (5.4−)5.6 − 6.6(−7.7) × (3.0−)3.2 − 4.1(−4.9) µm (Q = 1.42 − 1.85 µm, Q = 1.68 ± 0.13 µm), ellipsoid with blunt ends, uniseriate, hyaline, smooth-walled, 1 − 2 guttules. Asexual morph: Unknown.
Type: Southern Zambia, the Victoria Falls near Livingstone, on the ground mixed with putrid leaves and debris, 23 January 1981, Jiri Moravec (TRH:F:11406, isotype)
Material examined: China, Yunnan, Xishuangbanna, 101°25′ E, 21°41′ N, on soil, 27 August 2019, M. Zeng, ZM355 (HKAS 107301; MFLU 20-0257).
Notes: Originally, this was considered as a Phaedropezia species and named P. epispartius [Citation34,Citation35]. Acervus epispartius is the current name for the type species of Acervus. This species has cupulate to pulvinate apothecia with substipitate, small ascospores with guttules. In Jiri Moravec’s collection (TRH:F:11406), the size of ascospores are 6.5 − 7.6 × 3.8 − 4.9 µm, but we observed smaller ascospores in our collection (HMAS 107301). In addition, multiguttulate ascospores were found in collection TRH:F:11406, but we only found uniguttulate and biguttulate ascospores in our collection [Citation10].
Acervus globulosus Ekanayaka, Q. Zhao & K.D. Hyde, in Ekanayaka, Zhao, Jones, Pu & Hyde, Phytotaxa 283(1): 78 (2016) ()
Figure 3. Acervus globulosus (MFLU 20-0258). (a–f) Typical mature specimens. (g, h) Receptacle surface of pileus. (i) Asci and paraphyses. (j–l) Asci (l Asci in Melzer’s reagent.). (m–p) Apexes of asci (m–o Apexes in Congo red.). (q–u) Ascospores. Scale bars: c = 1500 μm. d–f = 1000 μm. g–l = 30 μm. m–u = 5 μm.

Index Fungorum No: IF 552369; Facesoffungi No: FoF 02524
Description: Saprobic on soil. Sexual morph: Apothecia 5 − 10 mm broad, 1 − 2 mm high, scattered to gregarious, arising from a yellowish mycelia pad, sessile, discoid to concave, hymenium yellow, receptacle surface orange to yellow. Medullary excipulum 138 − 270 µm broad, of textura angularis to textura globulosa, hyaline to yellowish, composed of 15 − 24 × 12 − 20 µm broad cells. Ectal excipulum 51 − 106 µm broad, of textura angularis, mix with textura epidermoidea, yellowish, composed of 18 − 23 × 13 − 18 μm broad, J-. Paraphyses 4 − 5 µm, yellow, unbranched, septate, filiform. Asci 82 − 100 × 6 − 8 µm, 8-spored, suboperculate, subcylindrical to clavate, J-. Ascospores [20/1/1, in H2O] (5.2−)5.4 − 6.1(−6.3) × (3.0−)3.3 − 4.0(−4.3) µm (Q = 1.41 − 1.78 µm, Q = 1.59 ± 0.18 µm), ellipsoid, uniseriate, hyaline, smooth-walled, 1 − 2 guttules. Asexual morph: Unknown.
Type: China, Yunnan, Xishuangbanna Botanical Garden, Menglun, Mengla County, 600 m alt., 101016′06.87ʺ E, 21055′11.69ʺ N, on soil near the vegetations of Millettia leptobtrya, Garcinia cowa, Castonopsis indica, 5 July 2014, W. Gang (HKAS 88987, holotype).
Material examined: Thailand, Chiang Mai, Samoeng, 98° 36′ E, 18° 56′ N, on soil, 13 July 2017, Y.P. Xiao, MM10 (MFLU 20-0258, HKAS 97493).
Notes: Ekanayaka et al. [Citation3] introduced this species which is distinguished by discoid to concave apothecia, textura angularis to textura globulosa cells in the excipulum, and ellipsoid ascospores. We observed some differences in our collection (MFLU 20-0258) of A. globulosus (). This species is the first recorded in Thailand.
Table 4. Morphological differences for two collections of Acervus globulosus.
Acervus rufus M. Zeng, Q. Zhao & K.D. Hyde, sp. nov. ()
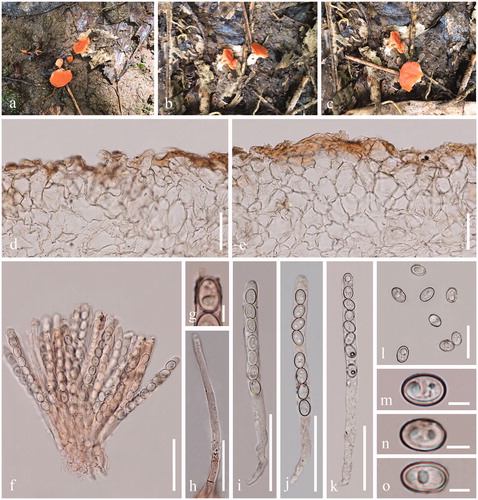
Figure 4. Acervus rufus (HKAS 107302, holotype). (a–c) Typic mature specimens. (d, e) Receptacle surface of pileus. f Hymenium in Congo red. (g) Apex of asci in Congo red. (h) Paraphyses in Congo red. (i–k) Asci (j Asci in Congo red. k Asci in Melzer’s reagent.). (l–o) Ascospores. Scale bars: d–f, i–k = 50 μm. h = 30 μm l = 20 μm. g, m–o = 5 μm.
Index Fungorum No: IF 557490; Facesoffungi No: FoF 07739
Etymology: The epithet refers to its reddish pileus and receptacle.
Description: Saprobic on soil with debris of decayed leaves. Sexual morph: Apothecia 7 − 10 mm broad, 8 − 10 mm high, scattered, arising from a white mycelia pad, forming a short stalked, 5 − 7 mm long, up to 5 mm broad, white to reddish. Receptacle discoid, red, hymenium red, receptacle surface reddish to orange. Medullary excipulum 87 − 163 µm broad, of textura angularis to textura globulosa mix with rarely swollen elongated hyphae, hyaline to reddish, composed of 19 − 30 × 12 − 18 µm broad cells. Ectal excipulum 127 − 201 µm broad, of textura angularis to textura epidermoidea, hyaline to reddish, 27 − 41 × 20 − 27 μm broad. Paraphyses 4 − 6 µm diam, hyaline, branched or unbranched, septate, filiform. Asci 121 − 140 × 8 − 9 µm, 8-spored, suboperculate to operculate, subcylindrical to clavate, J-. Ascospores [20/1/1, in H2O] (8.5−)9.1 − 10.6(−11.3) × (5.9−)6.5 − 7.6(−8.0) µm (Q = 1.20 − 1.59 µm, Q = 1.41 ± 0.12 µm), broadly ellipsoid, uniseriate, hyaline, smooth-walled, with 1 − 2 guttules. Asexual morph: Unknown.
Material examined: China, Xishuangbanna, Yunnan, 100°47′ E, 21°59′ N, on soil, 9 June 2018, M. Zeng, Zeng 012 (HKAS 107302, holotype); ibid. (MFLU 20-0259, isotype).
Notes: This species is characterized by red apothecia with short stalks, textura angularis cells in the excipulum, operculate asci and ellipsoid ascospores with 1 − 2 guttules. Based on phylogenetic tree, A. rufus groups with the A. flavidus, but A. flavidus differs in lemon yellow apothecia, sessile, medullary excipulum cells of textura intricata and 1 − 3-guttulate ascospores [Citation1,Citation3].
Acervus stipitatus Ekanayaka, Q. Zhao & K.D. Hyde, in Ekanayaka, Zhao, Jones, Pu & Hyde, Phytotaxa 283(1): 78 (2016) ()
Figure 5. Acervus stipitatus (HKAS 107302). (a) Habitat. (b, c) Typic mature specimens. (d) Receptacle surface of pileus. (e–g) Paraphyses (g Paraphyses in Congo red.). (h–j) Asci (i-i Asci in Congo red.). (k–m) Apexes of asci. (n–q) Ascospores. Scale bars: d, h–j = 30 μm. e–g = 20 μm. k–m, o–q = 5 μm. n = 15 μm.

Index Fungorum No: IF 552370; Facesoffungi No: FoF 02525
Description: Saprobic on soil. Sexual morph: Apothecia 0.7 − 1.0 cm broad, 1 − 2 mm high superficial, scattered, arising from a subhyaline to pale yellow mycelia pad, substipitate, discoid to concave, orange to yellow, hymenium yellowlish, receptacle yellow. Medullary excipulum 107 − 168 µm broad, of textura angularis to textura globulosa, hyaline to yellowish, composed of 19 − 25 × 13 − 17 µm broad cells. Ectal excipulum 106 − 217 µm broad, of textura angularis to textura epidermoidea, hyaline to yellowish, 21 − 28 × 15 − 19 μm broad, J-. Paraphyses 3 − 5 µm, hyaline to yellowish, branched, septate, filiform. Asci 89 − 100 × 5 − 7 µm, 8-spored, suboperculate to operculate, subcylindrical to clavate, J-. Ascospores [20/1/1, in H2O] (8.3−)8.9 − 11.0(−11.6) × (6.1−)6.4 − 7.8(−8.2) µm (Q = 1.08 − 1.62 µm, Q = 1.41 ± 0.13 µm), broadly ellipsoid, uniseriate, hyaline, smooth-walled with 1 to 2 or more small oil drops. Asexual morph: Unknown.
Type: China, Yunnan Province, Xishuangbanna, buffer zone of the Nabanhe National Nature Reserve (NNNR), 100032′–100044′ E, 22004′–22017′ N, 30 August 2015, S.C. Karunarathna (MFLU 16–0607, holotype).
Material examined: China, Yunnan, Xishuangbanna, 100°47′ E, 21°59′ N, on soil in a cave, 9 June 2018; M. Zeng, Zeng 013 (HKAS 107302, MFLU 20-0260).
Notes: Acervus stipitatus is characterized by discoid to concave apothecia with substipitate, uniguttulate to multiguttulate ascospores. A. stipitatus was established by Ekanayka et al. [Citation3]. In her descriptions, it is saprobic on dead stem with aseptate paraphyses, inoperculate asci, and uniguttulate ascospores. We reexamined holotype of A. stipitatus (MFLU 16-0607) and illustration is provided herein (). The A. stipitatus (MFLU 16-0607) has apothecia in which aseptate, unbranched, and hyaline paraphyses, operculate asci, and 1 to 3 guttules. The apothecial photo of the loaned herbarium differs from the previously published one [Citation3].
4. Discussion
The diversity of fungi in northern Thailand and southern China is proving to be extremely high [Citation36,Citation37]. Herein, we introduce one new species of Acervus and new collections from Yunnan Province and Thailand. Most Acervus species have been found on soil, though some species have also been reported from rotten wood, including A. flavidus, A. stipitatus, and A. xishuangbannicus, suggesting a saprobic lifestyle [Citation1,Citation3,Citation11]. Among all known Acervus species, nine have been reported in China including seven type species [Citation1–3,Citation11]. This indicates a high diversity of Acervus in China [Citation1,Citation3].
Acervus is a small group in the family Pyronemataceae [Citation16]. Phylogenetic methods have resolved some classification problems in this genus [Citation1–3,Citation13,Citation14]. Nonetheless, some features of Acervus remain problematic. For instance, ascus dehiscence, and whether asci are inoperculate, suboperculate or operculate are topics of debate [Citation4,Citation6,Citation9,Citation12]. In their description of A. beijingensis and A. changchunensis, Zhuang et al. [Citation1] stated that the “apical apparatus was not clearly seen”. Ren and Zhuang [Citation2] considered A. heilongjiangensis as an operculate species, but authors still stated, “apical apparatus not clearly seen under light microscope”. Thus, it is necessary to make additional collections of this species. Although Pezizomycetes are operculate discomycetes, the apical apparatus of some taxa in the class is in doubt [Citation12]. Thus, it is important to understand the apical apparatus for each group in this class. Regarding Acervus, Ekanayaka et al. [Citation3] considered the genus as an inoperculate discomycetes group. Nonetheless, an examination of our collections indicated distinct suboperculate to operculate structures for species in this genus.
Pfister [Citation9] provided a detailed explanation of A. epispartius as an older epithet and current name for the type in this genus. These observations were later confirmed by others [Citation1,Citation2,Citation10,Citation11]. Ekanayaka et al. [Citation3] treated A. epispartius and A. aurantiacus as separate without examining the holotype. We also did not observe the holotype of A. epispartius and A. aurantiacus. Given the disagreement, reexamination of holotype is recommended.
Herein a key to Acervus species is provided [Citation1–3,Citation10,Citation11]. Shape and size of ascospores are important characters to identify species [Citation1,Citation2]. In descriptions of Ren and Zhuang [Citation2], the size of ascospores is the primary morphological feature for distinguishing species. Up to now, the yellow to orange color of apothecia was considered an important trait to identify this genus. However, the new species A. rufus shows distinct red apothecia, thus expanding the range of apothecia colors in the genus. All species in the key have available sequence data, with the exception of A. xishuangbannicus and A. lusakianus. Therefore, future collections should focus on obtaining these species along with others to uncover the diversity of the genus. 
Acknowledgments
The authors thank Yuanpin Xiao (School of Science, Mae Fah Luang University, Thailand) for collecting samples. The authors thank the curator of Herbarium of Mae Fah Luang University for allowing us to examine the herbarium specimens.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Zhuang WY, Luo J, Zhao P. Two new species of Acervus (Pezizales) with a key to species of the genus. Mycologia. 2011;103(2):400–406.
- Ren F, Zhuang WY. A new species of Acervus (Pezizales) from China. Mycosystema. 2015;34:978–982.
- Ekanayaka AH, Zhao Q, Jones EBG, et al. Two novel Acervus species extend their distribution within Yunnan. China. Phytotaxa. 2016;283(1):74–82.
- Kanouse BB. Notes on new or unusual Michigan Discomycetes. V Mich Acad. 1938;23:149–154.
- Seaver FJ. The North American Cup-fungi (Inoperculates). The North American cup-fungi (Inoperculates). Lancaster, PA: Lancaster Press. 1951. p. 428.
- Korf RP. Discomycete flora of Asia. Precursor II: A revision of the genera Acervus and Ascosparassis and their new position in the Pezizales. Lloydia. 1963;26:21–26.
- Korf RP. Synoptic key to the genera of the Pezizales. Mycologia. 1972;64(5):937–994.
- Denison WC. Differentiation of tribes and genera in the family Sarcoscyphaceae. Persoonia. 1972;6:433–438.
- Pfister DH. The genus Acervus (Ascomycetes, Pezizales). Occas Pap Farlow Herb Cryptog Bot. 1975;8:1–11.
- Moravec J. Several operculate discomycetes from central and east Africa. Czech Mycol. 1983;37:237–251.
- Zhuang WY, Wang Z. Discomycetes of tropical China. II. Collections from Yunnan. Mycotaxon. 1998;69:339–358.
- Eckblad FE. The genera of operculate Discomycetes. A reevaluation of their taxonomy, phylogeny and nomenclature. Nord J Bot. 1968;15:1–191.
- Perry BA, Hansen K, Pfister DH. A phylogenetic overview of the family Pyronemataceae (Ascomycota, Pezizales). Mycol Res. 2007;111(Pt 5):549–571.
- Ekanayaka AH, Hyde KD, Jones EBG, et al. Taxonomy and phylogeny of operculate discomycetes: pezizomycetes. Fungal Divers. 2018;90(1):161–243.
- Wijayawardene NN, Hyde KD, Lumbsch HT, et al. Outline of Ascomycota: 2017. Fungal Divers. 2018;88(1):167–263.
- Wijayawardene NN, Hyde KD, Al-Ani LKT, et al. Outline of Fungi and fungus-like taxa. Mycosphere. 2020;11(1):1060–1456.
- Jayasiri SC, Hyde KD, Ariyawansa HA, et al. The Faces of Fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 2015;74(1):3–18.
- Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172(8):4238–4246.
- Rehner SA, Buckley E. A Beauveria phylogeny inferred from nuclear ITS and EF1-alpha sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 2005;97(1):84–98.
- Liu YJ, Whelen S, Hall BD. Phylogenetic relationships among Ascomycetes: evidence from an RNA polymerse II subunit. Mol Biol Evol. 1999;16(12):1799–1808.
- Matheny PB, Liu YJ, Ammirati JF, et al. Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales). Am J Bot. 2002;89(4):688–698.
- White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. 1990;18:315–322.
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98.
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780.
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25(15):1972–1973.
- Silvestro D, Michalak I. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 2012;12(4):335–337.
- Ronquist F, Teslenko M, Van Der Mark P, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542.
- Nylander JA, Ronquist JP, Huelsenbeck F, et al. Bayesian phylogenetic analysis of combined data. Syst Biol. 2004;53(1):47–67.
- Nuin PAS. Mtgui – a simple interface to model test. Program distributed by the author. University of Toronto. 2005. Available from: http://www.genedrift.org/
- Posada D, Buckley TR. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol. 2004;53(5):793–808.
- Rannala B, Yang Z. Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J Mol Evol. 1996;43(3):304–311.
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755.
- Cai L, Jeewon R, Hyde KD. Phylogenetic evaluation and taxonomic revision of Schizothecium based on ribosomal DNA and protein coding genes. Fungal Divers. 2005;19:1–21.
- Le Gal M. Les Discomycetes de Madagascar. Prodrome à une Flore Mycologique de Madagascar et Dépendances. Vol. 4. Paris, France: Académie des Sciences; 1953. p. 465.
- Le Gal M. Discomycetes du Congo belge d'apres les recoltes de Madame Goossens-Fontana. Bulletin du Jardin Botanique de L'État a Bruxelles. 1959;29(2):73–132.
- Hyde KD, Norphanphoun C, Chen J, et al. Thailand’s amazing diversity – up to 96% of fungi in northern Thailand are novel. Fungal Divers. 2018;93(1):215–239.
- Hyde KD, Dong Y, Phookamsak R, et al. Fungal diversity notes 1151–1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2020;100(1):5–277.