Abstract
Leaf spot on lemon balm is frequently observed in Korea, causing considerable damage to crops. In 2014 and 2015, the occurrence of leaf spot was observed in several production greenhouses at Suwon, Gongju, and Namwon in Korea. Symptoms on lower leaves initially developed as small, distinct, discolored lesions, which enlarged progressively turning into dark brown, angular spots surrounded by purplish-brown margins. Based on the morphological characteristics and sequence analysis of actin (ACT), translation elongation factor 1-alpha (EF-1α), internal transcribed spacer (ITS), 28S nrDNA (LSU), and RNA polymerase II second largest subunit (RPB2), the fungus associated with the lemon balm leaf spot was determined as Septoria melissae. To the best of our knowledge, this is the first report of lemon balm leaf spot caused by S. melissae in Asia as well as in Korea.
1. Introduction
Lemon balm (Melissa officinalis L.) is a perennial herb belonging to the family Lamiaceae, which occurs widely in central and southern Europe and Asia Minor. It is a well-known medicinal and culinary plant that is grown commercially in many parts of the world, including Korea. In Iran, it is used in traditional medicine for the treatment of headaches, flatulence, indigestion, colic, nausea, nervousness, anemia, vertigo, syncope, malaise, asthma, bronchitis, amenorrhea, cardiac failure, arrhythmias, insomnia, epilepsy, depression, psychosis, hysteria, ulcers, and wounds [Citation1].
Around 14 fungal isolates have been listed as being associated with various diseases of lemon balm worldwide. Several fungi cause leaf spot diseases, e.g. Phoma exigua var. exigua, Phyllosticta decidua, Pseudocercospora velutinomaculans, and Septoria melissae in Armenia and Europe [Citation2].
Septoria causing leaf spot disease on various plants, was first described in 1847 by Desmazières on the basis of Septoria cytisi [Citation3] and is a large genus with 2506 legitimate species [Citation4]. It was extremely complicated to classify Septoria into the correct species using traditional methods based on morphology because of the limitation of clear morphological characteristics and a significant number of variations [Citation3,Citation5]. The shape of fungi, including the genus Septoria, is affected by the environment, but DNA sequences are not [Citation5]. Many phylogenetic researches based on DNA sequences have been conducted to get an alternative resolution for the fungal taxonomy [Citation5–13]. It has been shown that multigene sequence analyses provided better correlation the DNA-based phylogenies with the morphological traits allowing accuracy on species identification.
This study aimed to investigate the occurrence and damage caused by leaf spot disease on lemon balm and determine the identity of the pathogens through morphological features, cultural characteristics, and sequencing approaches. This study will also contribute to the morphological and molecular Characterization of Septoria species.
2. Materials and methods
2.1. Occurrence of leaf spot
In February 2014, a leaf spot was first observed on pot-grown lemon balm in a plastic greenhouse in Suwon (37°16′21″N; 126°59′09″E), Korea. Similar leaf spot symptoms were also found in Gongju (36°29′41″N; 127°02′02″E) and Namwon (35°25′19″N; 127°31′23″E), Korea in 2014 and 2015. We have investigated the incidence of leaf spot, symptoms, and crop damage in Gongju and Namwon, which are significant lemon balm cultivation areas.
2.2. Single spore isolation
To obtain a single spore, conidiomata formed on the diseased leaf infected with leaf spots on the farm were identified with a loupe. The infected leaves were collected and stored for 3 days after sealing the samples in a plastic bag. White to cream-colored cirrhi was formed from conidiomata. Conidia were harvested from cirrhi with a drop of sterile water and then directly streaked using a bacterial loop onto 2% water agar media supplemented with 100 mg/L streptomycin sulfate. After 3 days of incubation at 25 °C, single conidial colonies were transferred to potato dextrose agar (PDA) with a sterile needle under a dissecting microscope.
2.3. Cultural and morphological observations
Morphological and colony characteristics of 2-week-old PDA single-spore isolates were used to determine colony characteristics and color while fungal structures from fresh samples mounted on a glass slide with a drop of water were observed using bright field and differential interference contrast light microscopy using an Olympus BX51 microscope (Olympus, Tokyo, Japan) for measurements. A Zeiss AX10 microscope equipped with an AxioCam MRc5 (Carl Zeiss, Göttingen, Germany) for imaging. Thirty measurements were taken at 400× and 1000× magnification for each sample.
2.4. Molecular phylogenetic analysis
To confirm the initial identification, a DNeasy Plant Mini Kit (Qiagen Inc., Valencia, CA) was used to extract genomic DNA from mycelia harvested from PDA cultures (KACC47728, JBARES80, JBARES81). Genomic DNA was extracted from 200 to 400 mg of fungal mycelia harvested from 2 weeks PDA cultures grown at 25 °C using a DNeasy Plant Mini Kit (Qiagen Inc., Valencia, CA). A sterile blade was used to scrape the mycelia from the surface of the culture plate. Five nuclear gene regions were targeted for PCR amplification and subsequent sequencing [Citation5]. The part of the actin gene (ACT) was amplified using the primer set ACT512F [Citation14] and ACT2Rd [Citation10], and region of the translation elongation factor 1-α gene (EF) using the primer set EF728F [Citation14] and EF-2 [Citation15]. The primer ITS5 or ITS1 and ITS4 [Citation16] were used to amplify the internal transcribed spacer areas as well as the 5.8S rRNA gene (ITS) of the rDNA. The primers part 28S nrDNA gene (LSU) was amplified using the primer set LSU1Fd and LR5 primers[Citation17], and part of the RNA polymerase II second largest subunit gene (RPB2) was amplified using the primer set fRPB2-5F and fRPB2-414R [Citation18], respectively. All PCR reaction mixtures and conditions followed those outlined by Hunter et al. [Citation19]. The amplicons were sequenced in both directions using the same PCR primers that were used for the initial amplification according to the manufacturer’s recommendations. The reactions were monitored using BigDye Terminator Cycle Sequencing Kits (Applied Biosystems, Foster City, CA) as indicated by the manufacturer and analyzed on an ABI 3130 automated DNA sequencer (Applied Biosystems).
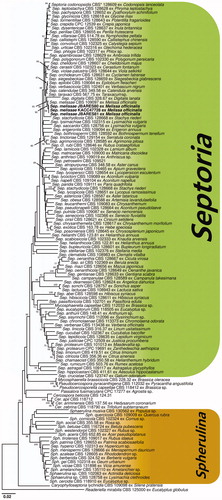
The possible identity of the isolates was established by comparing their actin (ACT), translation elongation factor 1-alpha (EF-1α), internal transcribed spacer (ITS), 28S nrDNA (LSU) and RNA polymerase II second largest subunit (RPB2) sequences with those in the GenBank database (National Center for Biotechnology Information [NCBI] US National Institute of Health, Bethesda, MD; http://www.ncbi.nlm.nih.gov/BLAST). Selected Septoria spp., Cercospora sp., and Mycosphaerella sp. sequences, including ACT, EF-1α, ITS, LSU, and RPB2, were retrieved from GenBank for the phylogenetic analysis. These retrieved sequences were included in the Septoria phylogenetic tree constructed by Verkley et al. [Citation5] and Quaedvlieg et al. [Citation3]. The obtained sequences were edited and assembled using the SeqMan software (Lasergene, DNASTAR, Madison, WI). A neighbor-joining phylogenetic tree was constructed using the maximum composite likelihood method by MEGA7 [Citation20]. The robustness of the NJ tree was evaluated with 1000 bootstraps (BS) values.
2.5. Pathogenicity test
Pathogenicity was tested by spraying 20 leaves of three-potted lemon balm with a conidial suspension (2 × 105 conidia/ml) harvested from a 4-week-old PDA culture. Ten control leaves were sprayed with sterile distilled water. The plants were placed in a dew chamber at 25 °C in darkness and continuous dew for the first 24 h and then moved to a greenhouse bench (25 ± 2 °C, 80% relative humidity).
3. Results
3.1. Incidence and damage of leaf spot
Disease incidence was determined at approximately 70%, while severity among those plants was found to be 30% (of leaves) prior to harvest resulting in considerable economic loss. The disease was more severe in greenhouse production than in open fields. Symptoms on lower leaves initially developed as small, distinct, and discolored lesions, which enlarged progressively turning into dark brown, angular, necrotic spots surrounded by purplish-brown margins (. The damage of plants caused by leaf spots was detracting from the marketable appearance of the plant, causing a decline in quality and yield. According to farmers who grow lemon balm, leaf spot occurs every year, and it is recognized as the main factor that significantly decreases yield.
Figure 1. Leaf spot of lemon balm (Melissa officinalis) infected with Septoria melissae. (A, B) Heavy infections detracting from the beauty of the plant; (C) Close-up symptoms in the later stage of disease development. Note the pycnidial conidiomata showing as small black dots on the lesions; (D) Conidiomata; (E) White cream-colored cirrhi of conidia being extruded through the ostioles of pycnidial conidiomata; (F) Conidia mass; (G) Conidia; (H) Two-week-old colonies of Septoria melissae growing on potato dextrose agar. Note plentiful production of conidia on the hazy gray and black colonies.

3.2. Isolation and accession of Septoria melissae
Three monoconidial isolates were prepared from white cream-colored cirrhi of conidia being extruded through the ostioles of pycnidial conidiomata formed on leaf spot affected leaves. One isolate came from Gongju in February 2015 and two isolates from infected leaves collected from the lemon balm farm of Namwon in September the same year. All isolates displayed similar colony morphology. A representative monoconidial isolate was deposited with the Korean Agricultural Culture Collection, Rural Development Administration, Wanju, Korea (accession no. KACC47728). The remaining isolates were lodged locally with the herbarium at Jeollabuk-do Agricultural Research and Extension Services (accession numbers JBARES80 and JBARES81). Samples collected were used for morphological Characterization and molecular analysis and submitted to the Korea University Herbarium (KUS; accession nos. KUS-F27803 and KUS-F27806).
3.3. Morphological characteristics of Septoria melissae
Characterization of colony morphology for all isolates was performed after 2 weeks of incubation on PDA at 25 °C with a 12-h photoperiod. The fungal colonies formed a slightly ruffled and mostly colorless margin; with a relatively small (5–7 mm diam.) colonies restricted to spreading, somewhat elevated in the center, the surface black in color, covered by a diffuse to the dense mat of finely felted, mostly gray aerial mycelium ().
Black conidiomata became visible on leaf lesions, and these were pycnidial, mostly epigenous, occasionally hypogenous, scattered to partly aggregated (usually two conidiomata closely adjacent), dark brown to rusty brown, globose, embedded in the host tissue or partly erumpent, 30–150 µm diam (). Ostioles were subcircular to irregular, 15–30 µm wide, surrounded with darker cells. White cream-colored cirrhi of conidia was extruded through the ostioles of pycnidial conidiomata (). Conidia were filiform, substraight to more or less curved, attenuated gradually to a narrowly rounded apex, attenuated gradually to an obconically subtruncate to rounded base, hyaline, with one to several oil drops per cell, 32–64 × 2.0–2.6 µm, and 2–5 septate (). Based on the morphological characteristics and host genus, the fungus was consistent with Septoria melissae Desm [Citation5,Citation21,Citation22].
3.4. Phylogenetic analysis of Septoria melissae
The PCR products of the three isolates of S. melissae by electrophoresis produced similar position banding, including ACT, EF-1α, ITS, LSU, and RPB2. The resulting 712 bp ACT, 512,547 bp ITS, 943 bp LSU, and 379 bp RPB2 sequences obtained from KACC47728 were deposited in GenBank (accession Nos. MH359403, MH359400, MH359402, MH359404, and MH359401, respectively).
A GenBank BLAST search was conducted with the ACT, EF-1α, ITS, LSU, and RPB2 sequences showing >99% identity with several Septoria sequences. Especially, as a result of a GenBank BLAST search for each gene regions of S. melissae, the ACT sequence shared 99% identity with KF253779, the EF-1α sequence shared 100% identity with KF253423, and the ITS sequence shared 100% identity with KF251475 and KJ683739, and the RPB2 sequence shared 100% identity with KF252472, respectively. However, LUS sequences have shown 100% identity with several Septoria sequences including S. melissae (KF251975), S. glycinicola (KF251934), S. cerastii (KF251866), S. digitalis (KF251905), respectively. S. glycinicola, S. cerastii, and S. digitalis are not reported in lemon barm globally, also their morphological characteristics were different from the Korean isolates.
A phylogenetic tree inferred from the ACT sequences indicates that our isolates were in the same group with S. melissae (KF253779). Similarly, the phylogenetic trees inferred from the EF-1α sequences indicates that our isolates were in the same group with S. melissae (KF253423), from the ITS sequences (KF251475) and from the LSU sequences (KF251979). Based on the nucleotide sequencs of the LSU region, the phylogenetic tree grouped together with numbers registered as S. glycinicola, S. cerastii, S. digitalis, S. putrida, S. bothriospermi, S. chrysanthemella, and S. lycopersici. A phylogenetic tree inferred from the RPB2 sequences indicates that our isolates were in the same group with S. melissae (KF252472). A phylogenetic tree based on the neighbor-joining (NJ) method inferred from the ACT, EF-1α, ITS, LSU, and RPB2 sequences showed that the isolates from our study formed a well-supported sister clade to S. melissae, and revealed a separate clade distinct from other species in the Septoria ().
Figure 2. Neighbor-joining trees based on Actin (ACT), translation elongation factor 1-alpha (EF-1α), internal transcribed spacer (ITS), 28S nrDNA (LSU), and RNA polymerase II second largest subunit (RPB2) sequences of Septoria melissae from lemon balm, and Septoria spp. retrieved from GenBank. The numbers above the nodes are the bootstrap values obtained from 1,000 replicates. The isolates obtained in this study are shown in boldface. *CBS: CBS-KNAW Fungal Biodiversity Center, Utrecht, the Netherlands.
3.5. Pathogenicity test
Pathogenicity testing by spraying leaves with conidia harvested from PDA culture yielded leaf spot symptoms identical to those observed in the field after 10 days. No symptoms were observed on control plants. S. melissae was re-isolated from the lesions of inoculated plants, fulfilling Koch’s postulates. Pathogenicity tests were conducted twice and obtained similar results.
4. Discussion
In the list of plant diseases in Korea, 101 species of Septoria are recorded in field crops, woody, and herbaceous plants, vegetables, ornamental plants, weeds, and flowers (the Society of Plant Pathology). Of the 14 diseases recorded on lemon balm internationally, Septoria leaf spot has been reported in Armenia, the Netherlands, Poland, Romania, and Uzbekistan [Citation2]. To our knowledge, this is the first report of the lemon balm leaf spot caused by S. melissae in Korea. The identification of S. melissae on lemon balm in Asia can be considered as a harmful threat to this well-known herb.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Dastmalchi K, Dorman HD, Oinonen PP, et al. Chemical composition and in vitro antioxidative activity of a lemon balm (Melissa officinalis L.) extract. LWT - Food Sci Technol. 2008;41(3):391–400.
- Farr DF, Rossman AY. Fungal databases, systematic mycology & microbiology laboratory. ARS, USDA [cited 2020 Mar 28]. Available from: http://nt.ars-grin.gov/fungaldatabases/.
- Quaedvlieg W, Verkley GJM, Shin HD, et al. Sizing up Septoria. Stud Mycol. 2013;75(1):307–390.
- Bakhshi M, Arzanlou M, Zare R, et al. New species of Septoria associated with leaf spot diseases in Iran. Mycologia. 2019;111(6):1056–1071.
- Verkley GJM, Quaedvlieg W, Shin HD, et al. A new approach to species delimitation in Septoria. Stud Mycol. 2013;75(1):213–305.
- Crous PW, Kang JC, Braun U. A phylogenetic redefinition of anamorph genera in Mycosphaerella based on ITS rDNA sequence and morphology. Mycologia. 2001;93(6):1081–1101.
- Crous P, Summerell B, Carnegie A, et al. Unravelling Mycosphaerella: do you believe in genera? Persoonia. 2009;23:99–118.
- Verkley G, Starink-Willemse M. A phylogenetic study of some Septoria species pathogenic to Asteraceae based on ITS ribosomal DNA sequences. Mycol Progress. 2004;3(4):315–323.
- Verkley G, Starink-Willemse M, van Iperen A, et al. Phylogenetic analyses of Septoria species based on the ITS and LSU-D2 regions of nuclear ribosomal DNA. Mycologia. 2004;96(3):558–571.
- Groenewald JZ, Nakashima C, Nishikawa J, et al. Species concepts in Cercospora: spotting the weeds among the roses. Stud Mycol. 2013;75(1):115–170.
- Golmohammadi R, Babai-Ahari A, Arzanlou M, et al. Morphological and molecular identification and pathogenicity of Septoria species involved in leaf spot disease on Populus species in East Azerbaijan, West Azerbaijan and Ardabil provinces. Iran J Plant Pathol. 2015;51(3):357–365.
- Bakhshi M, Arzanlou M. Multigene phylogeny reveals a new species and novel records and hosts in the genus Ramularia from Iran. Mycol Progress. 2017;16(7):703–712.
- Bakhshi M, Arzanlou M. Multigene phylogeny and morphotaxonony of Septoria spp. from Iran along with a checklist of Septoria-like taxa. Rostaniha. 2018;18:122–141.
- Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91(3):553–556.
- O'Donnell K, Kistler HC, Cigelnik E, et al. Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc Natl Acad Sci USA. 1998;95(5):2044–2049.
- White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocol. 1990;18:315–322.
- Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172(8):4238–4246.
- Quaedvlieg W, Kema GHJ, Groenewald JZ, et al. Zymoseptoria gen. nov.: a new genus to accommodate Septoria-like species occurring on graminicolous hosts. Persoonia. 2011;26:57–69.
- Hunter GC, Wingfield BD, Crous PW, et al. A multi-gene phylogeny for species of Mycosphaerella occurring on Eucalyptus leaves. Stud Mycol. 2006;55:147–161.
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874.
- Rădulescu E, Negru A, Docea E. Septoriozele din România: Editura Academiei Republicii Socialiste România; 1973.
- Vanev SG, Sameva EF, Bakalova GG. Order Sphaeropsidales. Fungi Bulgaricae. 1997;3:1–335.
