Abstract
The genus Pholiota (Strophariaceae, Basidiomycota) is made up of wood-rotting saprotrophic mushrooms characterized by a yellow or brown pileus with scales and/or slimy, and by a brownish smooth spore with a germ pore. However, these features are not enough to distinguish its species, or separate the genus Pholiota from other brown-spored wood-rotting genera such as Hypholoma and Stropharia. Although internal transcribed spacer (ITS) sequence-based identification has improved identification accuracy for species of Pholiota, most Pholiota species in Korea are reported based on morphological features. To evaluate the taxonomy of Pholiota species, we investigated 62 specimens collected from 1999 to 2019 in Korea using ITS sequence analysis and morphological observation. Twelve of the 16 recorded Pholiota species in Korea were identified. While eight species were clearly separated, the ITS analysis did not distinguish three in the Pholiota adiposa complex. Therefore, further investigation is required to distinguish these three species. ITS sequences deposited in GenBank confirm that P. highlandensis exists in Korea. The presence of the other four Pholiota species could not be confirmed through specimens or sequence information in GenBank. A taxonomic key and the ITS sequence data for Korean Pholiota species are included and can be good baselines for further research on Pholiota taxonomy and diversity.
1. Introduction
The genus Pholiota Kummer (1871) is composed of saprotrophic flesh mushrooms in the family Strophariaceae [Citation1]. Pholiota is characterized by a yellow or brown pileus with scales, brownish smooth surface spore, and brown spore print [Citation2,Citation3]. Pholiota squarrosa is designated as a type species [Citation2]. According to the current overview of Basidiomycota [Citation4], approximately 157 species are recorded in this genus. Pholiota species are commonly found in temperate climate regions and they perform important roles in the ecosystem as wood decomposers and soil saprotrophs [Citation2,Citation3].
Pholiota species produce a variety of bioactive compounds that can have antitumor and antioxidant effects [Citation5,Citation6]. Activity and application of lignocellulase from P. adiposa have been reported in several studies [Citation7,Citation8]. Some Pholiota species are edible—e.g. P. microspora is well-known for its culinary usage in Asian countries [Citation2,Citation9], whereas P. squarrosa is poisonous [Citation10].
The presence or absence of pleurocystidia and cheilocystidia, cystidial incrustation, wall thickness, and coloration have been used as key characteristics to distinguish between Pholiota species [Citation2,Citation3]. However, morphological characters are not enough to distinguish the species because macro-morphological characteristics of Pholiota species are variable depending on the environmental conditions, and micro-morphological characteristics are often very similar between species. For example, a gelatinous layer can be detected from the fruiting body only during the fresh state, and some species show the gelatinous characteristic only when mature, so it is difficult to identify them when they are collected as immature basidiocarps. Moreover, morphological characteristics can be diverse, even within the same species [Citation2,Citation3]. As such, it is important to proceed with further identification using molecular analysis, which has become an increasingly important tool for accurately identifying fungal species [Citation11,Citation12]. We recently discovered many new fungal species and amended misidentified species by reevaluating the other genera using molecular analysis [Citation13–19].
Phylogenetic studies have placed Pholiota species within the family Strophariaceae, but they form a paraphyletic clade with Hypholoma and Stropharia species [Citation20–23]. Recent phylogenetic analysis based on the internal transcribed spacer (ITS) sequence has improved the accuracy at which Pholiota species are identified and has led to the recognition of new Pholiota species [Citation24–27].
Eighteen species of Pholiota have been reported in Korea over the years, but only 16 of these are currently accepted [Citation28–30]. Since P. adiposa and P. squarrosa were first reported in Korea in 1940 [Citation31], 13 additional Pholiota species were reported based on morphology by 2011. Through molecular analysis, P. alnicola was transferred to genus Flammula in Hymenogastraceae [Citation32], and its name changed to Flammula alnicola (Fr.) P. Kumm. [Citation33]. Recently, P. abietis, P. multicingulata, and P. limonella were additionally reported by phylogenic analysis using ITS sequence [Citation29,Citation30,Citation34]. Later, P. abietis was confirmed to be synonymous with P. limonella [Citation35]. Most Pholiota species were mainly identified based on the features of their basidiocarp, so it is necessary to re-evaluate Pholiota based on molecular analysis. In this study, we investigate the species diversity of Pholiota in Korea based on ITS sequence analysis and morphology.
2. Material and methods
2.1. Sample collection and observation
A total of 62 Pholiota specimens were obtained from three herbaria in South Korea: 12 from the National Institute of Biological Resources (NIBR), 28 from the Korea National Arboretum (KA), and 22 from the Seoul National University Fungal Collections (SFC). All samples were collected from 1999 to 2019 in South Korea () and were stored dried. Pictures of the fresh fruiting bodies and information on the collection location and date were available, but there was often no accurate ecological data. To observe the microscopic features, the specimens were mounted in 5% (w/v) KOH and 5% (w/v) Congo red solution, and then were observed using an Eclipse 80i light microscope (Nikon, Tokyo, Japan). At least 30 basidiospores, 10 basidia, 10 cheilocystidia, and 10 pleurocystidia were measured per specimen.
Table 1. Summary and GenBank accession numbers for Pholiota specimens used in this study.
2.2. DNA extraction, PCR amplification, and sequencing
Genomic DNA was extracted from the fruiting bodies using a modified cetyltrimethylammonium bromide (CTAB) extraction protocol [Citation36]. The ITS region was amplified using primers (ITS1F/ITS4B) that target the ITS region [Citation37]. PCR amplifications were performed on a thermal cycler (C1000TM; Bio-Rad, Richmond, CA) using the AccuPower PCR premix (Bioneer Co., Daejeon, Korea). The PCR conditions were 95 °C for 5 min; followed by 35 cycles of 95 °C for 40 sec, 55 °C for 40 sec, and 72 °C for 1 min; and finally 72 °C for 5 min. PCR products were loaded on to a 1% agarose gel and purified using the ExpinTM PCR Purification Kit (GeneAll Biotechnology, Seoul, Korea). Samples were sequenced by Sanger sequencing using the aforementioned primers at Macrogen (Seoul, Korea) using an ABI PRISM 3730XL Analyzer (Applied Biosystems, Foster City, CA).
2.3. Phylogenetic analyses
ITS sequences of each sample were proofread using MEGA7 [Citation38] and deposited in GenBank (accession numbers in ). Sequences were aligned using the Multiple Alignment Fast Fourier Transform (MAFFT ver. 7) [Citation39] with ITS sequences of Pholiota obtained from GenBank and UNITE. The alignments were checked manually, and upon verification, ambiguous alignments were adjusted. A Neighbor Joining (NJ) Tree was constructed also using MEGA7 with 1000 bootstraps [Citation38]. Agrocybe species were selected as outgroups, in accordance with previous studies [Citation22,Citation23].
3. Results
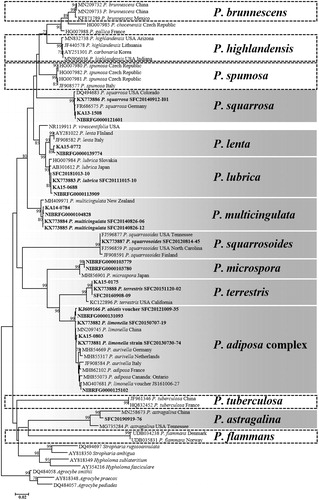
A total of 62 specimens were divided into nine groups based on the ITS analysis. Each group included 1–23 specimens (). Representative sequences from each group were selected, and phylogenetic analysis was performed together with ITS sequences downloaded from GenBank and UNITE. The nine groups were (): Pholiota adiposa complex (number of specimens = 23), P. astragalina (n = 1), P. lenta (n = 2), P. lubrica (n = 8), P. microspora (n = 2), P. multicingulata (n = 11), P. squarrosa (n = 6), P. squarrosoides (n = 1), and P. terrestris (n = 8). The P. adiposa complex includes morphologically similar species: P. adiposa, P. aurivella, and P. limonella. Although P. highlandensis was not present in our analysis, the ITS sequence of P. carbonaria (accession number: AY251301) deposited from Korea is identical to that of P. highlandensis.
Figure 1. Phylogenetic trees based on neighbor-joining (NJ) analysis of the ITS region in Pholiota species. Bootstrap support values (1000 replicates) >70% are presented. The recorded Korean Pholiota species are placed in boxes. The shaded boxes represent confirmed species and the dotted boxes represent unidentified species in Korea. The scale bar indicates the number of nucleotide substitutions per site.
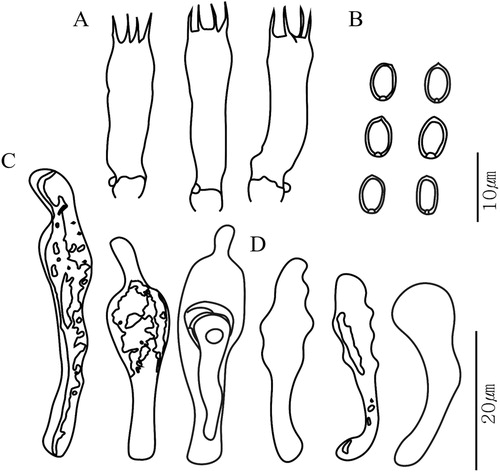
Pictures of each species are shown in . Basidiospores, basidia, and cystidia were well observed from the dried specimens, and their size and shape were in good agreement with previous reports. Basidiospores were generally elliptical, thick walled with an apical pore, and ranged from 4–9 µm long and 2–6 µm wide. Basidia were clavate in shape and 12–33 µm long. Types of cystidia observed were cheilocystidia, pleurocystidia, and caulocystidia. Cheilocystidia and pleurocystidia were observed in all species except P. microspora. Caulocystidia was only observed in P. squarrosoides and P. terrestris in our specimens. Chrysocystidia were detected in the species of the P. adiposa complex, P. squarrosa, P. squarrosoides, and P. terrestris. The microscopic features of the type species P. squarrosa are shown in .
4. Taxonomic key for Korean Pholiota
1. Pileus cuticle lacking any gelatinous layers (surface granulose to fibrillose or scaly, or rarely canescent, glabrous, and hygrophanous) P. squarrosa
1. Pileus cuticle with a layer of gelatinized hyphae in some part (surface may be glabrous to scaly) 2
2. Pileus typically distinctly scaly at the time the veil breaks; chrysocystidia or somewhat similar structures present in hymenium3
2. Pileus fibrillose, scaly or granulose, lacking chrysocystidia in hymenium7
3. Gelatinous subcutis evident only with maturity; pileus surface initially densely covered with dry squamules4
3. Gelatinous subcutis evident throughout development; pileus surface initially viscid and glabrous or more sparsely covered with squamules than above 5
4. Pileus dark grayish brown to dark cinnamon, stipe with scales colored like those on pileus P. terrestris
4. Pileus ground color pallid, inner veil white P. squarrosoides
5. Strongly glutinous pileus covered with a thick layer of hyaline slime, in the absence of any cystidia P. microspora
5. Pileus glutinous in moist weather, presence of cheilocystidia 6
6. Spore length shorter than 5 µm P. flammans
6. Spore length longer than 5 µm P. adiposa complex
7. Pleurocystidia none. The pileus generally appears dry and appressed fibrillose to echinate‐squamulose P. tuberculosa
7. Pleurocystidia present and prominently projecting 8
8. Always fruiting on burned ground around charcoal 9
8. Habitat typically lignicolous, more rarely on soil or humus 10
9. Stipe 1.5–4 (5) mm thick; veil pallid at first P. highlandensis
9. Stipe 5–10 (5) mm thick; pileus dark yellow‐brown; veil lemon yellow young P. brunnescens
10. Spores 7–10 × 4–6 µm 11
10. Spores smaller 5–7.5 (8) × 3–4.5 (5) µm 12
11. Pileus 1–3.5 cm wide, stipe 1–3.5 mm thick P. multicingulata
11. Pileus wider than 3.5 cm, and stipe thicker than 3.5 mm P. spumosa
12. Taste bitter and black discoloration P. astragalina
12. Taste mild to farinaceous 13
13. Pileus cinnamon to dark cinnamon brown or bay‐brown; marginal area not yellowP. lubrica
13. Pileus variously colored, marginal area pallid to grayish and developing yellow tones in ageP. lenta
5. Discussion
Pholiota species are often confused with the members of Stropharia and Hypholoma. Species across the three genera can be distinguished by the color of their spore prints. Pholiota species have basidiospores that are dark gray-brown to dark ocher-brown or dark reddish-brown without violet or purple tones, while the latter two have violet- to purple-brown or violet- to purple-black basidiospores [Citation40]. In addition to the discernible color of the spores, the presence of scales on a yellowish cap is also a general characteristic that differentiates Pholiota from Stropharia and Hypholoma. However, as the three genera share too many overlapping morphological characteristics and form a paraphyly [Citation20–23], further research is needed to distinguish their relationships.
We identified 12 of the 16 recorded Pholiota species from Korea in this study. Eight species were clearly separated from the ITS tree. On the other hand, three species that grouped into the P. adiposa complex were not distinguished by the ITS analysis. Many mycologists acknowledge the complexity around distinguishing these three species because they share many morphological features [Citation2,Citation35]. Their genetic similarities have also been proven from several other studies. Matsumoto et al. [Citation41] reported that these three species clustered together in an RFLP analysis of ITS, large subunit rDNA, and intergenic spacer (IGS). Papp and Dima [Citation42] grouped P. adiposa, P. limonella, and P. cerifera into the P. adiposa complex as they formed a monophyletic group and was not distinguished by ITS sequence analysis.
The key distinguishable features of P. adiposa, P. aurivella, and P. limonella are the size of the basidiospores and their host preference [Citation2,Citation35,Citation43]. P. limonella has slightly smaller but distinctly narrower spores than do the other two species [Citation43]. The spore size of P. limonella is 6.5–9 × 4–5.3 µm, while those of P. adiposa and P. aurivella are 7.5–9.5 × 5–6.2 µm and 7.5–10.5 × 5–6.5 µm, respectively. In addition, P. aurivella only resides on Salix, while P. limonella prefers Betula, and P. adiposa is found on various deciduous trees, and sometimes even on conifers [Citation43–45]. However, it seems that these differences may be due to environmental or intraspecific variation. In this study, we did not have enough ecological information or consistent ITS sequences for the three species in the P. adiposa complex to distinguish them. To determine whether these three species are of the same or different species, it is necessary to conduct more detailed morphological observations and mating tests, assess their ecological preferences, and compare other genetic markers.
While the ectomycorrhizal species composition in Korea is very different from those of Europe and North America [Citation16,Citation17], there is little difference between saprotrophic fungi compositions between continents [Citation46,Citation47]. Correspondingly, we confirmed that the Korean Pholiota species showed little genetic difference from the European and North American Pholiota species in the ITS neighbor-joining (NJ) phylogeny (). Pholiota species seem to be distributed over a wide area, which may explain the low genetic variance across continents.
Our specimens did not include five of the previously reported Pholiota species (P. brunnescens, P. flammans, P. highlandensis, P. spumosa, and P. tuberculosa) in Korea. However, a P. carbonaria (accession number: AY251301) of Korean origin was deposited in GenBank, and was identified as P. highlandensis, a pyrophilous species that is synonymous with P. carbonaria [Citation48]. Regarding P. brunnescens, there is an environmental sequence (accession number: LC100010) deposited from Japan [Citation48], and P. spumosa [Citation27] and P. tuberculosa (GenBank accession number: JF961346) have been reported from China. Therefore, it is highly possible that these three species also exist in Korea. The presence of P. flammans could not be confirmed through specimens, nor through DNA sequence information in any open Database.
In conclusion, we confirmed 12 species of Pholiota from Korea based on morphological and sequence analyses. Further investigation is required to distinguish the three species associated with the P. adiposa complex. Identification of species in this genus requires a comprehensive consideration of morphological and molecular characteristics. Identification using a BLAST search of ITS sequences has recently become popular because sequencing has become affordable and the available sequences in databases have increased. However, it should be noted that there are inaccurate sequences in the databases [Citation49–51]. Our research may serve as a good baseline for the study of the taxonomy and the diversity of Pholiota in Korea to discover new species and to investigate the ecological roles of Pholiota. The taxonomic key for Pholiota in Korea is presented based on external references to compensate for the lack of ecological data. This will be useful for further identifying Pholiota species in Korea.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Kummer P. Der Führer in die Pilzkunde. Zerbst, Germany: C. Luppe; 1871.
- Smith AH, Hesler LR. The North American species of Pholiota. New York: Hafner Publishing Company; 1968.
- Holec J. The genus Pholiota in central and western Europe. Eching, Germany: IHW-Verlag und Verlagsbuchhandlung; 2001.
- He MQ, Zhao RL, Hyde KD, et al. Notes, outline and divergence times of Basidiomycota. Fungal Divers. 2019;99(1):105–367.
- Gan D, Ma L, Jiang C, et al. Production, preliminary characterization and antitumor activity in vitro of polysaccharides from the mycelium of Pholiota dinghuensis Bi. Carbohydr Polym. 2011;84(3):997–1003.
- Hu Q, Wang H, Ng TB. Isolation and purification of polysaccharides with anti-tumor activity from Pholiota adiposa (Batsch) P. Kumm. (higher Basidiomycetes). Int J Med Mushr. 2012;14(3):271–284.
- Dhiman SS, Jagtap SS, Jeya M, et al. Immobilization of Pholiota adiposa xylanase onto SiO2 nanoparticles and its application for production of xylooligosaccharides. Biotechnol Lett. 2012;34(7):1307–1313.
- Jagtap SS, Dhiman SS, Jeya M, et al. Saccharification of poplar biomass by using lignocellulases from Pholiota adiposa. Bioresour Technol. 2012;120:264–272.
- Neda H. Correct name for “nameko”. Mycoscience. 2008;49(1):88–91.
- Shaffer RL. Poisoning by Pholiota squarrosa. Mycologia. 1965;57(2):318–319.
- Schoch CL, Seifert KA, Huhndorf S, et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci USA. 2012;109(16):6241–6246.
- Hibbett D, Abarenkov K, Kõljalg U, et al. Sequence-based classification and identification of fungi. Mycologia. 2016;108(6):1049–1068.
- Min YJ, Park MS, Fong JJ, et al. Molecular taxonomical re-classification of the genus Suillus Micheli ex S. F. Gray in South Korea. Mycobiology. 2014;42(3):221–228.
- Park MS, Quan Y, Jung PE, et al. Re-evaluation of the genus Antrodia (Polyporales, Basidiomycota) in Korea. Mycobiology. 2014;42(2):114–119.
- Lee H, Park MS, Jung PE, et al. Re-evaluation of the taxonomy and diversity of Russula section Foetentinae (Russulales, Basidiomycota) in Korea. Mycoscience. 2017;58(5):351–360.
- Lee H, Wissitrassameewong K, Park MS, et al. Taxonomic revision of the genus Lactarius (Russulales, Basidiomycota) in Korea. Fungal Divers. 2019;95(1):275–335.
- Cho HJ, Park MS, Lee H, et al. A systematic revision of the ectomycorrhizal genus Laccaria from Korea. Mycologia. 2018;110(5):948–961.
- Jung PE, Lee H, Wu SH, et al. Revision of the taxonomic status of the genus Gloeoporus (Polyporales, Basidiomycota) reveals two new species. Mycol Prog. 2018;17(7):855–863.
- Park KH, Oh SY, Park MS, et al. Re-evaluation of Armillaria and Desarmillaria in South Korea based on ITS/tef1 sequences and morphological characteristics. For Path. 2018;48(6):e12447.
- Moncalvo JM, Vilgalys R, Redhead SA, et al. One hundred and seventeen clades of euagarics. Mol Phylogenet Evol. 2002;23(3):357–400.
- Walther G, Garnica S, Wei M. The systematic relevance of conidiogenesis modes in the gilled Agaricales. Mycol Res. 2005;109(Pt 5):525–544.
- Petersen G, Knudsen H, Seberg O. Alignment, clade robustness and fungal phylogenetics—Crepidotaceae and sister families revisited. Cladistics. 2010;26(1):62–71.
- Matheny PB, Moreau PA, Vizzini A, et al. Crassisporium and Romagnesiella: two new genera of dark-spored. Agaricales. Syst Biodivers. 2015;13(1):28–41.
- Holec J, Kolařík M. Pholiota gallica nom. nov., based on P. lubrica var. obscura. Mycotaxon. 2014a;127(1):161–171.
- Holec J, Kolařík M, Bizio E. Pholiota chocenensis—a new European species of section Spumosae (Basidiomycota, Strophariaceae). Mycol Prog. 2014b;13(2):399–406.
- Siegel N, Nguyen NH, Vellinga EC. Pholiota olivaceophylla, a forgotten name for a common snowbank fungus, and notes on Pholiota nubigena. Mycotaxon. 2015;130(2):517–532.
- Tian E, Bau T, Ding Y. A new species of Pholiota subgenus Flammuloides section Lubricae (Strophariaceae, Agaricales) from Tibet, China. Phytotaxa. 2016;286(3):153–160.
- Lee YS, Lim YW, Kim JJ, et al. National list of species of Korea: Basidiomycota. Incheon, Korea: National Institute of Biological Resources; 2015.
- Cho HJ, Lee H, Park JY, et al. Seven new recorded species in five genera of the strophariaceae in Korea. Mycobiology. 2016;44(3):137–145.
- Lee JS, Kim C, Choi S, et al. Eight previously unreported species of macrofungi from Korea. Kor J Med Mycol. 2017;45(4):362–369.
- Kaburagi Y. Korean and Manchurian practical manual of forest. Korea Forest Experiment Station. Tokyo: Yokendo; 1940. pp. 357–361.
- Matheny PB, Curtis JM, Hofstetter V, et al. Major clades of Agaricales: a multilocus phylogenetic overview. Mycologia. 2006;98(6):982–995.
- Redhead SA. Proposal to conserve the name Flammula (Fr.) P. Kumm. (Fungi: Agaricales) against Flammula (Webb ex Spach) Fourr. (Spermatophyta: Ranunculaceae). Taxon. 2013;62(2):401–402.
- Lee WD, Lee H, Fong JJ, et al. A checklist of the basidiomycetous macrofungi and a record of five new species from Mt. Oseo in Korea. Mycobiology. 2014;42(2):132–139.
- Farr ER, Miller OK, Jr, Farr DF. Biosystematic studies in the genus Pholiota, stirps Adiposa. Can J Bot. 1977;55(9):1167–1180.
- Rogers SO, Bendich AJ. Extraction of total cellular DNA from plants, algae and fungi. In: Gelvin SB, Schilperoort RA, editors. Plant molecular biology manual D1. Dordrecht: Springer; 1994. pp. 1–8.
- Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2(2):113–118.
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874.
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780.
- Breitenbach J, Kränzlin F. Fungi of Switzerland, Vol. 4. Agarics 2nd part, Entolomataceae, Pluteaceae, Amanitaceae, Agaricaceae, Coprinaceae, Bolbitaceae, Strophariaceae. Mycologia Luzern (Schweiz): Mykologia; 1995.
- Matsumoto T, Fukumasa-Nakai Y, Nagasawa E, et al. Phylogenetic position of Pholiota nameko in the genus Pholiota inferred from restriction analysis of ribosomal DNA. Mycoscience. 2003;44(3):197–202.
- Papp V, Dima B. A Pholiota squarrosoides első magyarországi előfordulása és előzetes filogenetikai vizsgálata [First record and preliminary ITS phylogeny of Pholiota squarrosoides from Hungary]. Mikol Közl, Clusiana. 2014;53(1–2):33–42.
- Holec J. The taxonomy of Pholiota aurivella and Pholiota adiposa—a return to Batsch and Fries. Czech Mycol. 1998;50(3):201–221.
- Jacobsson S. On the correct interpretation of Pholiota adiposa and a taxonomic survey of section Adiposae. Windahlia. 1987;17:1–18.
- Jacobsson S. Pholiota in northern Europe. Windahlia. 1990;19:1–86.
- Sato H, Tsujino R, Kurita K, et al. Modelling the global distribution of fungal species: new insights into microbial cosmopolitanism. Mol Ecol. 2012;21(22):5599–5612.
- Park JH, Pavlov IN, Kim MJ, et al. Investigating wood decaying fungi diversity in Central Siberia, Russia using ITS sequence analysis and interaction with host trees. Sustainability. 2020;12(6):2535–2535.
- Matheny PB, Swenie RA, Miller AN, et al. Revision of pyrophilous taxa of Pholiota described from North America reveals four species-P. brunnescens, P. castanea, P. highlandensis, and P. molesta. Mycologia. 2018;110(6):997–1016.
- Nilsson RH, Ryberg M, Kristiansson E, et al. Taxonomic reliability of DNA sequences in public sequence databases: a fungal perspective. PLoS One. 2006;1(1):e59.
- Jung PE, Fong JJ, Park MS, et al. Sequence validation for the identification of the white-rot fungi Bjerkandera in public sequence databases. J Microbiol Biotechnol. 2014;24(10):1313–1319.
- Jargalmaa S, Eimes JA, Park MS, et al. Taxonomic evaluation of selected Ganoderma species and database sequence validation. PeerJ. 2017;5:e3596.