Abstract
Species of the genus Aspergillus have a variety of effects on humans and have been considered industrial cell factories due to their prominent ability for manufacturing several products such as heterologous proteins, secondary metabolites, and organic acids. Scientists are trying to improve fungal strains and re-design metabolic processes through advanced genetic manipulation techniques and gene delivery systems to enhance their industrial efficiency and utility. In this review, we describe the current status of the genetic manipulation techniques and transformation methods for species of the genus Aspergillus. The host strains, selective markers, and experimental materials required for the genetic manipulation and fungal transformation are described in detail. Furthermore, the advantages and disadvantages of these techniques are described.
1. Introduction
Species of the genus Aspergillus are widely distributed among natural environments and have several effects on humans [Citation1,Citation2]. More than 300 species have been identified to date, of which several strains also have detrimental or beneficial effects for humans [Citation3]. For example, Aspergillus fumigatus and other human pathogenic Aspergillus species cause aspergillosis, including invasive aspergillosis, chronic pulmonary aspergillosis, and allergic bronchopulmonary aspergillosis [Citation4–7]. Several fungi (A. flavus, A. parasiticus, and A. ochraceus) infect agricultural crops during the harvest or post-harvest stages, and spoil crops or produce detrimental secondary metabolites in them, called mycotoxins, causing mycotoxin contamination [Citation8–11]. Although Aspergillus spp. have detrimental effects on humans, these species are also beneficial for the food and pharmaceutical industries [Citation12–14]. A. niger and A. oryzae serve as factories that produce the organic acids and enzymes that are required for various industries [Citation12,Citation14]. For the food industry, generally recognized as safe fungi are used in the preparation of traditional fermented foods [Citation15]. Many researchers are investigating the development of novel fungal strains, heterologous expression systems, and novel secondary metabolites through advanced genetic manipulation techniques because of the usefulness of these fungi.
Sequencing analyses of approximately 19 genomes of the species Aspergillus were performed, as well as several comparative genome analyses of various strains in the same species, and it proved to be very useful for understanding the function of genes and the species Aspergillus [Citation3]. Through genomic analyses, transcriptomic, proteomic, and metabolomic analyses have helped understand fungal biology and find useful genes or metabolic gene clusters [Citation16,Citation17]. Advanced genetic manipulation techniques are being developed and applied to fungal strain improvement and gene utilization in order to understand and use the functions of various useful genes [Citation18,Citation19]. However, unlike Escherichia coli or Saccharomyces cerevisiae, the species of Aspergillus have limited selective markers, low efficiency transformation tools, and constraint of genetic engineering tools. Therefore, advanced genetic manipulation techniques are being developed and applied to fungal strain improvement and gene utilization. Furthermore, genetic research and strain improvement are developing very quickly through genetic engineering techniques such as the clustered regularly-interspaced short palindromic repeats (CRISPR)–Cas system [Citation19,Citation20]. In this review, we present recent information on genetic modification, transformation, and selection techniques, and host strains required for genetic research in species of Aspergillus.
2. Host strains and selective markers
One of the most important items in the entire genetic engineering experiment is the selection of a host strain and a selective marker gene suitable for the genetic manipulation methods. Appropriate host strains and selective markers are essential for reducing the probability of false positive transformants and obtaining a high-yield final product.
2.1. Fungal host strains
Wild type strains can be used for genetic engineering in some fungal species because the genome information is available [Citation21]. However, modified strains are generally used to increase the efficiency of trials and experiments [Citation22]. For example, protease deficient strains are generally used in heterologous protein expression systems [Citation23–25]. The most important item to consider when selecting host strains is which auxotrophic selective marker, except for drug resistant markers, is used in the genetic engineering [Citation26–28]. This is because host strains with specific gene deletions should be used when an auxotrophic selective marker is used in genetic engineering. In most filamentous fungi, the pyrG, or pyrG ortholog genes, which encode for orotidine-5′-monophosphate decarboxylase, are used as auxotrophic markers; therefore, pyrG defect mutants are used for host strains [Citation27,Citation29–31]. Another important consideration is the selection of a host strain with high transformation efficiency. Homologous integration frequency was reportedly increased by Ku-deficient mutation in most filamentous fungi [Citation32–36].
2.2. Selective marker genes
In species of Aspergillus, 2 selective markers, drug resistance markers, and auxotrophic markers are widely used for genetic engineering experiments [Citation21,Citation37] (). Drug (antibiotic) resistance genes are generally used as dominant selection markers which are not necessary for special host strains [Citation38]. If one of the Aspergillus species is susceptible to an antibiotic, it can be widely used in any species. The most commonly used marker gene is a hygromycin resistance gene, which can be used in most filamentous fungi including A. fumigatus, A. oryzae, A. nidulans, and A. terreus [Citation38–40]. However, A. flavus NRRL 3357 is resistant to the hygromycin and cannot be used for transformation. Instead of hygromycin, the phleomycin and pyrithiamine resistance genes have been used for A. flavus transformation [Citation27]. With these genes, the bleomycin resistance gene has been used for drug resistance markers in other species of Aspergillus [Citation27,Citation41]. Although drug resistance markers have the advantage of not needing a specific host strain, their use is limited due to expensive antibiotics and issues with genetically modified organisms.
Table 1. Lists of certain selective markers and cell wall degrading enzymes used in Aspergillus transformation.
Auxotrophic genes, defined as genes that encode an essential protein for biosynthesis of an essential nutrient, are widely used for selection of positive transformants for use in genetic engineering [Citation42]. As mentioned above, the pyrG gene is widely used as a selective marker for Aspergillus transformation [Citation43]. Various pyrG-deficient mutants have been developed and these strains cannot grow in media without uridine or uracil, but they can survive by adding uridine or uracil to the medium, so it is easy to use in selection [Citation30,Citation44]. Another important feature of the pyrG marker is that it can be developed with a recyclable marker system, so multiple genes can be knocked out [Citation45–47]. Although the pyrG gene is widely used in various Aspergillus species, other auxotrophic marker genes such as argB and sC are used only with a few Aspergillus species [Citation37]. Several mutant strains were developed as the host strains in A. nidulans or A. oryzae in order to use the auxotrophic genes. For example, Oakley et al. [Citation33] developed a variety of auxotrophic heterologous markers that can be used in A. nidulans selection. In A. oryzae, other auxotrophic markers such as argB, niaD, and adeA have been used for selection markers for transformation [Citation37]. If multiple selective markers can be used in a host strain, it is very convenient and efficient to generate multiple deletion mutants or complemented strains [Citation28,Citation48].
3. Methods of fungal transformation
Unlike bacteria or yeast strains, transformation efficiency is low in filamentous fungi, and therefore the transformation process is considered as a bottleneck in genetic engineering [Citation21,Citation49]. Also, species of Aspergillus contain a rigid cell wall, which is the main hurdle for increasing transformation efficiency. Currently, several transformation methods have been developed for Aspergillus spp., and three methods, including polyethylene glycol (PEG)-mediated transformation (PMT), Agrobacterium-mediated transformation (AMT), and electroporation (EP), are mostly used. Only a few studies have been conducted though the biolistic transformation (BT) system in A. nidulans and A. giganteus [Citation50,Citation51]. Therefore, the process and strategies for these three methods will be discussed in below ().
Figure 1. Transformation methods for species of Aspergillus. (A) Procedures of PEG-mediated transformation (PMT); (B) Agrobacterium-mediated transformation (AMT); and (C) electroporation (EP) methods used in species of Aspergillus.
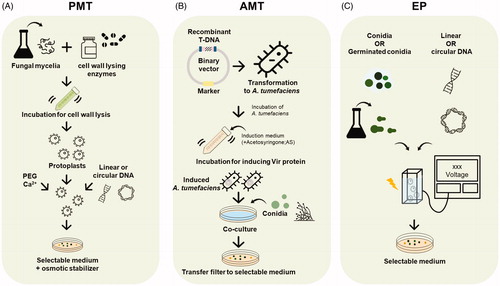
3.1. PEG‑mediated transformation
Among the various transformation methods, PMT is the most used method with species of Aspergillus. PMT was primarily explored in yeast [Citation52,Citation53]. PMT has used in A. nidulans for transferring the acetamidase-containing plasmid [Citation54]. The procedure described in this paper was later modified and applied to another species of Aspergillus. Then, Szewczyk et al. [Citation55] reported a well-organized PMT protocol, and the methods in this protocol were modified and used according the species.
The PMT procedure simply involves fungal culture, protoplast preparation via cell wall degradation, DNA delivery via PEG incubation, transformant regeneration, and selection (). Protoplasts are cells with the cell walls removed, which are mainly used as recipient cells for PMT [Citation56]. The process of generating protoplasts is a key step in PMT [Citation55]. Germ tubes or young mycelia are mainly used for Aspergillus protoplast transformation, treated with various cell wall lysing enzymes dependent on fungal species and cell types. For fungal cell wall lysis, yatalase (from Corynebacterium spp; consist of chitinase, chitobiase, and beta-1,3-glucanase; Takara), VinoTaste Pro (from Trichoderma harzianum and A. niger consist of polygalacturonase and 1,3-β-glucanase; Novozyme), and lysing enzymes (from T. harzianum, mixture of β-glucanase, cellulase, protease, and chitinase; Sigma-Aldrich) are mostly used as lysing enzymes, and they are mixed according to the species (). After protoplast generation, the protoplast solution is mixed with the exogenous DNA (PCR products or plasmid) under a high concentration of PEG (40–50%) and CaCl2 condition which induce DNA uptake and membrane permeability. Protoplasts are sensitive to osmotic stress, so during preparation and regeneration of protoplast osmotic stabilizers, such as potassium chloride and sorbitol, they should be added into the buffer solution and selective media.
Overall, PMT is widely used with many filamentous fungi because it does not require expensive equipment and the procedure of PMT is simple [Citation49,Citation55]. In addition, PMT is not necessary the special vector and bacteria required for AMT. However, in order to achieve success in transformation, culture condition, buffer composition, and cell wall degrading enzymes should be optimized for each species. For some species of Aspergillus, including A. nidulans, A. oryzae, and A. fumigatus, the procedure of PMT is well established [Citation37,Citation55,Citation57], but it is essential for optimize an appropriate method, especially lysing enzyme, for other species.
3.2. Agrobacterium‑mediated transformation
Agrobacterium tumefaciens is a plant pathogenic bacterium which causes crown gall disease in plants [Citation58]. This bacterium contains tumor-inducing plasmid (Ti-plasmid) which can enter the plant cell and insert of small transfer DNA (T-DNA) into the infected plant cell genome. Using this principle, A. tumefaciens is used as a vector for gene transfer in various filamentous fungi. AMT was first used for genetic transformation in species of Aspergillus in 1998 by de Groot et al. [Citation59]. Until now, AMT has been used for the exogenous DNA introduction in more than 10 fungal species (). Among these studies, Michielse et al. [Citation63] well described the detailed protocol in A. awamori.
Table 2. Lists of AMT in species of Aspergillus.
The AMT procedure includes construction of binary factor, transformation of A. tumefaciens with binary factor, preparation of fungal conidia, Agrobacterium-fungal cocultivation, and transformation selection (). AMT is different from PMT and its advantage is that it does not required protoplasts for host starter cell types [Citation49]. Some researchers have used protoplasts as starting materials, but in most studies, conidia or germinated conidia are mainly used for starting materials [Citation59,Citation63]. AMT also has several advantages such as high transformation frequency, high gene-replacement frequency, and single-copy T-DNA integration [Citation63]. As a result of conducting several transformation methods such as AMT, PMT, EP, or BT, AMT exerts high transformation efficiency and stability in A. giganteus and A. fumigatus [Citation51,Citation62]. However, unlike PMT, AMT requires the processes of construction of a binary vector, which containing a fungal selective marker and the gene of interest, and co-culturing A. tumefaciens and fungi, so it takes a longer time compared to other transformation methods. To make AMT a success, therefore, it is important to construct the appropriate vectors and optimize the culture condition, such as the ratio of Agrobacterium:conidia concentration, cocultivation condition, and acetosyringone concentration, to induce the vir gene [Citation63].
3.3. Electroporation
EP is a highly efficient method for introducing exogenous DNA into a cell by applying a high-voltage electric pulse [Citation76]. EP was used for the first time in A. nidulans, and at this time, protoplasts are being used for recipient cells of exogenous DNA [Citation77]. To increase efficiency and reduce time by removing the protoplast preparation process, several research groups used germinated conidia or conidia in Aspergillus spp. including A. oryzae, A. niger, and A. nidulans [Citation78–80] (). In addition, several factors should be optimized, including electric field intensity, pulse condition, DNA concentration, and buffer composition [Citation49]. Although EP is a simple and convenient method compared to other transformation methods, expensive instrumentation is required for EP (). Currently, EP is rarely used by researchers for fungal studies.
Table 3. Lists of EP-mediated transformation in species of Aspergillus.
4. Genetic manipulation tools
Genetic manipulation of filamentous fungi has been critical for understanding fungal biology and developing fungal species for industries. In particular, the identification and production of novel fungal metabolites through genetic manipulation have been used in the pharmaceutical and industrial fields. A variety of genetic manipulation tools have been developed in bacteria or mammalian systems and applied in fungal biology. Of these methods, two genome editing tools, such as homologous recombination (HR)-mediated gene targeting and CRISPR/Cas9 genome editing, are commonly used in fungal research, and they will be discussed below ().
Figure 2. Genome editing methods in species of Aspergillus. (A) Procedures of homologous recombination (HR)-mediated gene targeting method and (B) CRISPR-Cas system-mediated gene targeting method used in species of Aspergillus.
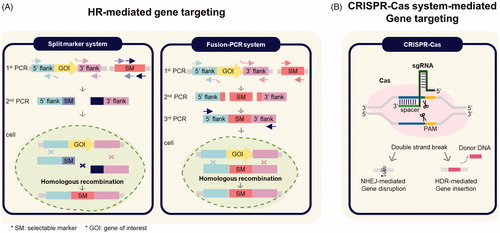
4.1. HR-mediated gene targeting
HR-mediated gene targeting is a powerful tool and is most often used for deleting interest of genes, inserting epitope tags, and replacing promoters in species of Aspergillus [Citation55]. This method can be used in many investigations on species of Aspergillus due to the high transformation efficiency of ku-deficient strain, the easy production of PCR-based products, and the optimization of transformation methods and selective markers. Especially, PCR-based cassette construction for HR has the advantage of shortening cloning time and increasing transformation efficiency than cloning by using the vector-based method.
To generate the PCR product for HR, two systems such as the split-marker and fusion (or joint) PCR systems are mainly used [Citation55,Citation85] (). The split-marker system requires 2 DNA products containing overlapping fragments of a selective marker such as pyrG or hph (hygB) [Citation86–88]. Two DNA fragments are introduced into the recipient strains and then these products replace the gene of interest via the HR events [Citation89]. The fusion (or joint) PCR system is used more than the split-marker system, and has the advantage of being used for gene tagging and promoter replacement [Citation55,Citation90]. The fusion PCR system generally requires 3 PCR products, 2 fragments for the HR in the genomic DNA flaking a selection region, and the other for a selective marker with or without the tags or the replaceable promoter. These 3 fragments are fused together by joint PCR generating a linear cassette suitable for transformation. The linear PCR cassettes made with the fusion PCR method are generally introduced through PMT methods into the recipient fungal cells, mainly ku-deficient strains. This procedure is the most common and widely used method of genetic engineering in species of Aspergillus. The use HR-mediated gene targeting is possible only when there are many available selective markers in the fungal species.
4.2. CRISPR–Cas system
In recent years, genome editing using the engineered or bacterial nucleases, such as zinc-finger nucleases, transcription activator-like effector nucleases, and CRISPR–Cas-associated nucleases, have been developed and widely used in almost all eukaryotic systems [Citation91]. Even though these nucleases are also used in filamentous fungi, the CRISPR–Cas system is the most widely used with many filamentous fungi [Citation19]. The CRISPR/Cas system was discovered in the prokaryotic organisms for adaptive immune system against foreign elements and a useful genome editing system has been developed for most eukaryotic systems [Citation92,Citation93]. The CRISPR/Cas system needs 2 components, the Cas9 nuclease and a single chimeric guide RNA (sgRNA) for gene editing in filamentous fungi [Citation19] (). The engineered sgRNA, consisting of the Cas-binding site and a spacer that recognizes a protospacer sequence in the specific target site, interacts with the Cas nuclease, which then forms the Cas-sgRNA complex. This complex binds to the target sequence and a short protospacer adjacent motif (PAM), which then make a DNA double-strand break (DSB) within the target DNA (3-5 upstream of the PAM). The DSB in the target site is then repaired by the self-repair mechanism pathways, non-homologous end joining (NHEJ) pathway or homology directed repair (HDR) pathway. The NHEJ pathway is an active repair system but can lead to nucleotide insertions or deletions at DSB site; thereby, it can disrupt the function of the targeted gene. Or, in the presence of donor DNA, the HDR pathway, high-fidelity but less efficient repair system, can insert the donor DNA at the DSB site.
The CRISPR/Cas system was first applied for genome editing system in several species of Aspergillus in 2015 [Citation20]. Nodvig et al. generated a vector for the CRISPR/Cas9 system called pFC332 which contains the hph gene, the codon-optimized Streptococcus pyogenes cas9 gene under tef1 promoter, and the sgRNA genes under the A. nidulans gpdA promoter. This vector was introduced through the PMT method into the fungal protoplast and then it disrupted a single target gene in several species of Aspergillus (). Based on this CRISPR/Cas system, several groups are currently studying fungal biology using this system and, in particular, it is used to develop industrially useful strains in A. niger and A. oryzae. The development and application of the CRISPR/Cas system will be an important cornerstone for the development of industrial strains and novel fungal metabolites in the future.
Table 4. Lists of the CRISPR/Cas system in species of Aspergillus.
5. Conclusions and perspectives
Species of the genus Aspergillus have been of great important fungi because of their beneficial and detrimental effects on human. Species of Aspergillus can produce various enzymes and organic acids, considering as cell factories. These fungi can produce many secondary metabolites, so it can be utilized as host species for generating novel metabolites that can be used for pharmaceutical purposes. Recent studies reported that species of Aspergillus can decompose plastics and pollutants, so these fungi can be utilized for the environmental field. Furthermore, through the development of genetic engineering and systemic biology, many researchers sought to use filamentous fungi as cell factories. Unlike bacteria or yeast, the available gene editing methods and transformation efficiency are low, but filamentous fungi are still attractive cell factories for industry. To develop Aspergillus as cell factories, currently, PMT and HR-mediated gene targeting are mainly used in genetic engineering. Recently, the CRISPR genome editing method was used for gene editing, but the use of other gene editing methods developed in bacteria and yeast is still marginal. Therefore, it is necessary to develop a more efficient transformation method and a system for genetic manipulation.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bennett JW. An overview of the genus Aspergillus. In: Machida M, Gomi K, editors. Aspergillus: molecular biology and genomics. Norfolk (UK): Caister Academic Press; 2010. p. 1–17.
- Samson RA, Visagie CM, Houbraken J, et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud Mycol. 2014;78:141–173.
- de Vries RP, Riley R, Wiebenga A, et al. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 2017;18:28.
- Latge JP. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev. 1999;12:310–350.
- Paulussen C, Hallsworth JE, Alvarez-Perez S, et al. Ecology of aspergillosis: insights into the pathogenic potency of Aspergillus fumigatus and some other Aspergillus species. Microb Biotechnol. 2017;10:296–322.
- Latge JP, Chamilos G. Aspergillus fumigatus and Aspergillosis in 2019. Clin Microbiol Rev. 2019;33:e00140-18.
- Bastos RW, Valero C, Silva LP, et al. Functional characterization of clinical isolates of the opportunistic fungal pathogen Aspergillus nidulans. mSphere. 2020;5:e00153-20.
- Hedayati MT, Pasqualotto AC, Warn PA, et al. Aspergillus flavus: human pathogen, allergen and mycotoxin producer. Microbiology (Reading). 2007;153:1677–1692.
- Perrone G, Susca A, Cozzi G, et al. Biodiversity of Aspergillus species in some important agricultural products. Stud Mycol. 2007;59:53–66.
- Perrone G, Gallo A. Aspergillus species and their associated mycotoxins. Methods Mol Biol. 2017;1542:33–49.
- Agriopoulou S, Stamatelopoulou E, Varzakas T. Advances in analysis and detection of major mycotoxins in foods. Foods. 2020;9:518.
- Kitamoto K. Cell biology of the Koji mold Aspergillus oryzae. Biosci Biotechnol Biochem. 2015;79:863–869.
- Park HS, Jun SC, Han KH, et al. Diversity, application, and synthetic biology of industrially important Aspergillus fungi. Adv Appl Microbiol. 2017;100:161–202.
- Cairns TC, Nai C, Meyer V. How a fungus shapes biotechnology: 100 years of Aspergillus niger research. Fungal Biol Biotechnol. 2018;5:13.
- Bourdichon F, Casaregola S, Farrokh C, et al. Food fermentations: microorganisms with technological beneficial use. Int J Food Microbiol. 2012;154:87–97.
- Lu H, Cao W, Liu X, et al. Multi-omics integrative analysis with genome-scale metabolic model simulation reveals global cellular adaptation of Aspergillus niger under industrial enzyme production condition. Sci Rep. 2018;8:14404.
- Ojeda-Lopez M, Chen W, Eagle CE, et al. Evolution of asexual and sexual reproduction in the aspergilli. Stud Mycol. 2018;91:37–59.
- Wang S, Chen H, Tang X, et al. Molecular tools for gene manipulation in filamentous fungi. Appl Microbiol Biotechnol. 2017;101:8063–8075.
- Song R, Zhai Q, Sun L, et al. CRISPR/Cas9 genome editing technology in filamentous fungi: progress and perspective. Appl Microbiol Biotechnol. 2019;103:6919–6932.
- Nodvig CS, Nielsen JB, Kogle ME, et al. A CRISPR-Cas9 system for genetic engineering of filamentous fungi. PLoS One. 2015;10:e0133085.
- Ruiz-Diez B. Strategies for the transformation of filamentous fungi. J Appl Microbiol. 2002;92:189–195.
- Balabanova LA, Shkryl YN, Slepchenko LV, et al. Development of host strains and vector system for an efficient genetic transformation of filamentous fungi. Plasmid. 2019;101:1–9.
- van den Hombergh JP, van de Vondervoort PJ, Fraissinet-Tachet L, et al. Aspergillus as a host for heterologous protein production: the problem of proteases. Trends Biotechnol. 1997;15:256–263.
- Yoon J, Maruyama J, Kitamoto K. Disruption of ten protease genes in the filamentous fungus Aspergillus oryzae highly improves production of heterologous proteins. Appl Microbiol Biotechnol. 2011;89:747–759.
- Xie H, Ma Q, Wei D, et al. Metabolic engineering of an industrial Aspergillus niger strain for itaconic acid production. 3 Biotech. 2020;10:113.
- Jin FJ, Maruyama J, Juvvadi PR, et al. Development of a novel quadruple auxotrophic host transformation system by argB gene disruption using adeA gene and exploiting adenine auxotrophy in Aspergillus oryzae. FEMS Microbiol Lett. 2004;239:79–85.
- He ZM, Price MS, Obrian GR, et al. Improved protocols for functional analysis in the pathogenic fungus Aspergillus flavus. BMC Microbiol. 2007;7:104.
- Niu J, Arentshorst M, Seelinger F, et al. A set of isogenic auxotrophic strains for constructing multiple gene deletion mutants and parasexual crossings in Aspergillus niger. Arch Microbiol. 2016;198:861–868.
- Palmer LM, Cove DJ. Pyrimidine biosynthesis in Aspergillus nidulans: isolation and preliminary characterisation of auxotrophic mutants. Mol Gen Genet. 1975;138:243–255.
- Xue T, Nguyen CK, Romans A, et al. Isogenic auxotrophic mutant strains in the Aspergillus fumigatus genome reference strain AF293. Arch Microbiol. 2004;182:346–353.
- Nguyen KT, Ho QN, Pham TH, et al. The construction and use of versatile binary vectors carrying pyrG auxotrophic marker and fluorescent reporter genes for Agrobacterium-mediated transformation of Aspergillus oryzae. World J Microbiol Biotechnol. 2016;32:204.
- da Silva Ferreira ME, Kress MR, Savoldi M, et al. The akuB(KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot Cell. 2006;5:207–211.
- Nayak T, Szewczyk E, Oakley CE, et al. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics. 2006;172:1557–1566.
- Takahashi T, Masuda T, Koyama Y. Enhanced gene targeting frequency in ku70 and ku80 disruption mutants of Aspergillus sojae and Aspergillus oryzae. Mol Genet Genomics. 2006;275:460–470.
- Meyer V, Arentshorst M, El-Ghezal A, et al. Highly efficient gene targeting in the Aspergillus niger kusA mutant. J Biotechnol. 2007;128:770–775.
- Chang PK, Scharfenstein LL, Wei Q, et al. Development and refinement of a high-efficiency gene-targeting system for Aspergillus flavus. J Microbiol Methods. 2010;81:240–246.
- He B, Tu Y, Jiang C, et al. Functional genomics of Aspergillus oryzae: strategies and progress. Microorganisms. 2019;7:103.
- Gravelat FN, Askew DS, Sheppard DC. Targeted gene deletion in Aspergillus fumigatus using the hygromycin-resistance split-marker approach. Methods Mol Biol. 2012;845:119–130.
- Punt PJ, Oliver RP, Dingemanse MA, et al. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene. 1987;56:117–124.
- Ventura L, Ramon D. Transformation of Aspergillus terreus with the hygromycin B resistance marker from Escherichia coli. FEMS Microbiol Lett. 1991;66:189–193.
- Suzuki S, Tada S, Fukuoka M, et al. A novel transformation system using a bleomycin resistance marker with chemosensitizers for Aspergillus oryzae. Biochem Biophys Res Commun. 2009;383:42–47.
- Pronk JT. Auxotrophic yeast strains in fundamental and applied research. Appl Environ Microbiol. 2002;68:2095–2100.
- Nguyen KT, Ho QN, Do L, et al. A new and efficient approach for construction of uridine/uracil auxotrophic mutants in the filamentous fungus Aspergillus oryzae using Agrobacterium tumefaciens-mediated transformation. World J Microbiol Biotechnol. 2017;33:107.
- Oakley BR, Rinehart JE, Mitchell BL, et al. Cloning, mapping and molecular analysis of the pyrG (orotidine-5'-phosphate decarboxylase) gene of Aspergillus nidulans. Gene. 1987;61:385–399.
- Nielsen ML, Albertsen L, Lettier G, et al. Efficient PCR-based gene targeting with a recyclable marker for Aspergillus nidulans. Fungal Genet Biol. 2006;43:54–64.
- Maruyama J, Kitamoto K. Multiple gene disruptions by marker recycling with highly efficient gene-targeting background (DeltaligD) in Aspergillus oryzae. Biotechnol Lett. 2008;30:1811–1817.
- Tani S, Tsuji A, Kunitake E, et al. Reversible impairment of the ku80 gene by a recyclable marker in Aspergillus aculeatus. AMB Express. 2013;3:4.
- Dohn JW Jr, Grubbs AW, Oakley CE, et al. New multi-marker strains and complementing genes for Aspergillus nidulans molecular biology. Fungal Genet Biol. 2018;111:1–6.
- Li D, Tang Y, Lin J, et al. Methods for genetic transformation of filamentous fungi. Microb Cell Fact. 2017;16:168.
- Herzog RW, Daniell H, Singh NK, et al. A comparative study on the transformation of Aspergillus nidulans by microprojectile bombardment of conidia and a more conventional procedure using protoplasts treated with polyethyleneglycol. Appl Microbiol Biotechnol. 1996;45:333–337.
- Meyer V, Mueller D, Strowig T, et al. Comparison of different transformation methods for Aspergillus giganteus. Curr Genet. 2003;43:371–377.
- Hutchison HT, Hartwell LH. Macromolecule synthesis in yeast spheroplasts. J Bacteriol. 1967;94:1697–1705.
- Anne J, Eyssen H, Somer PD. Formation and regeneration of Penicillium chrysogenum protoplasts. Arch Microbiol. 1974;98:159–166.
- Tilburn J, Scazzocchio C, Taylor GG, et al. Transformation by integration in Aspergillus nidulans. Gene. 1983;26:205–221.
- Szewczyk E, Nayak T, Oakley CE, et al. Fusion PCR and gene targeting in Aspergillus nidulans. Nat Protoc. 2006;1:3111–3120.
- Peberdy JF. 1995. Fungal protoplasts. In: Kück U, editor. Genetics and biotechnology. The mycota (a comprehensive treatise on fungi as experimental systems for basic and applied research). Berlin (Germany): Springer. p. 49–60.
- Zhao C, Fraczek MG, Dineen L, et al. High-throughput gene replacement in Aspergillus fumigatus. Curr Protoc Microbiol. 2019;54:e88.
- Gelvin SB. Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol Mol Biol Rev. 2003;67:16–37.
- de Groot MJ, Bundock P, Hooykaas PJ, et al. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat Biotechnol. 1998;16:839–842.
- Gouka RJ, Gerk C, Hooykaas PJ, et al. Transformation of Aspergillus awamori by Agrobacterium tumefaciens-mediated homologous recombination. Nat Biotechnol. 1999;17:598–601.
- Park S-M. Improved transformation of the filamentous fungus Aspergillus niger using Agrobacterium tumefaciens. Mycobiology. 2001;29:132–134.
- Sugui JA, Chang YC, Kwon-Chung KJ. Agrobacterium tumefaciens-mediated transformation of Aspergillus fumigatus: an efficient tool for insertional mutagenesis and targeted gene disruption. Appl Environ Microbiol. 2005;71:1798–1802.
- Michielse CB, Hooykaas PJ, van den Hondel CA, et al. Agrobacterium-mediated transformation of the filamentous fungus Aspergillus awamori. Nat Protoc. 2008;3:1671–1678.
- Kunitake E, Tani S, Sumitani J, et al. Agrobacterium tumefaciens-mediated transformation of Aspergillus aculeatus for insertional mutagenesis. AMB Express. 2011;1:46.
- Li M, Zhou L, Liu M, et al. Construction of an engineering strain producing high yields of α-transglucosidase via Agrobacterium tumefaciens-mediated transformation of Asperillus niger. Biosci Biotechnol Biochem. 2013;77:1860–1866.
- Kalleda N, Naorem A, Manchikatla RV. Targeting fungal genes by diced siRNAs: a rapid tool to decipher gene function in Aspergillus nidulans. PLoS One. 2013;8:e75443.
- Mora-Lugo R, Zimmermann J, Rizk AM, et al. Development of a transformation system for Aspergillus sojae based on the Agrobacterium tumefaciens-mediated approach. BMC Microbiol. 2014;14:247.
- Wang D, He D, Li G, et al. An efficient tool for random insertional mutagenesis: Agrobacterium tumefaciens-mediated transformation of the filamentous fungus Aspergillus terreus. J Microbiol Methods. 2014;98:114–118.
- Fan Z, Yu H, Guo Q, et al. Identification and characterization of an anti-oxidative stress-associated mutant of Aspergillus fumigatus transformed by Agrobacterium tumefaciens. Mol Med Rep. 2016;13:2367–2376.
- Weyda I, Yang L, Vang J, et al. A comparison of Agrobacterium-mediated transformation and protoplast-mediated transformation with CRISPR-Cas9 and bipartite gene targeting substrates, as effective gene targeting tools for Aspergillus carbonarius. J Microbiol Methods. 2017;135:26–34.
- Han G, Shao Q, Li C, et al. An efficient Agrobacterium-mediated transformation method for aflatoxin generation fungus Aspergillus flavus. J Microbiol. 2018;56:356–364.
- Min T, Xiong L, Liang Y, et al. Disruption of stcA blocks sterigmatocystin biosynthesis and improves echinocandin B production in Aspergillus delacroxii. World J Microbiol Biotechnol. 2019;35:109.
- Sun Y, Niu Y, He B, et al. A dual selection marker transformation system using Agrobacterium tumefaciens for the industrial Aspergillus oryzae 3.042. J Microbiol Biotechnol. 2019;29:230–234.
- Setoguchi S, Mizutani O, Yamada O, et al. Effect of pepA deletion and overexpression in Aspergillus luchuensis on sweet potato shochu brewing. J Biosci Bioeng. 2019;128:456–462.
- Zhu SY, Xu Y, Yu XW. Improved homologous expression of the acidic lipase from Aspergillus niger. J Microbiol Biotechnol. 2020;30:196–205.
- Chakraborty BN, Kapoor M. Transformation of filamentous fungi by electroporation. Nucleic Acids Res. 1990;18:6737.
- Richey MG, Marek ET, Schardl CL, et al. Transformation of filamentous fungi with plasmid DNA by electroporation. Phytopathology. 1989;79:844–847.
- Chakraborty BN, Patterson NA, Kapoor M. An electroporation-based system for high-efficiency transformation of germinated conidia of filamentous fungi. Can J Microbiol. 1991;37:858–863.
- Ozeki K, Kyoya F, Hizume K, et al. Transformation of intact Aspergillus niger by electroporation. Biosci Biotechnol Biochem. 1994;58:2224–2227.
- Sanchez O, Aguirre J. Efficient transformation of Aspergillus nidulans by electroporation of germinated conidia. Fungal Genet Newsl. 1996;43:48–51.
- Brown JS, Aufauvre-Brown A, Holden DW. Insertional mutagenesis of Aspergillus fumigatus. Mol Gen Genet. 1998;259:327–335.
- Weidner G, d'Enfert C, Koch A, et al. Development of a homologous transformation system for the human pathogenic fungus Aspergillus fumigatus based on the pyrG gene encoding orotidine 5'-monophosphate decarboxylase. Curr Genet. 1998;33:378–385.
- Firon A, Beauvais A, Latge JP, et al. Characterization of essential genes by parasexual genetics in the human fungal pathogen Aspergillus fumigatus: impact of genomic rearrangements associated with electroporation of DNA. Genetics. 2002;161:1077–1087.
- Firon A, Villalba F, Beffa R, et al. Identification of essential genes in the human fungal pathogen Aspergillus fumigatus by transposon mutagenesis. Eukaryot Cell. 2003;2:247–255.
- Kuck U, Hoff B. New tools for the genetic manipulation of filamentous fungi. Appl Microbiol Biotechnol. 2010;86:51–62.
- Sheppard DC, Doedt T, Chiang LY, et al. The Aspergillus fumigatus StuA protein governs the up-regulation of a discrete transcriptional program during the acquisition of developmental competence. Mol Biol Cell. 2005;16:5866–5879.
- Nielsen ML, de Jongh WA, Meijer SL, et al. Transient marker system for iterative gene targeting of a prototrophic fungus. Appl Environ Microbiol. 2007;73:7240–7245.
- Nielsen JB, Nielsen ML, Mortensen UH. Transient disruption of non-homologous end-joining facilitates targeted genome manipulations in the filamentous fungus Aspergillus nidulans. Fungal Genet Biol. 2008;45:165–170.
- Goswami RS. Targeted gene replacement in fungi using a split-marker approach. Methods Mol Biol. 2012;835:255–269.
- Yu JH, Hamari Z, Han KH, et al. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol. 2004;41:973–981.
- Gaj T, Sirk SJ, Shui SL, et al. Genome-editing technologies: principles and applications. Cold Spring Harb Perspect Biol. 2016;8:a023754.
- Barrangou R, Marraffini LA. CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol Cell. 2014;54:234–244.
- Rath D, Amlinger L, Rath A, et al. The CRISPR-Cas immune system: biology, mechanisms and applications. Biochimie. 2015;117:119–128.
- Fuller KK, Chen S, Loros JJ, et al. Development of the CRISPR/Cas9 system for targeted gene disruption in Aspergillus fumigatus. Eukaryot Cell. 2015;14:1073–1080.
- Zhang C, Meng X, Wei X, et al. Highly efficient CRISPR mutagenesis by microhomology-mediated end joining in Aspergillus fumigatus. Fungal Genet Biol. 2016;86:47–57.
- Katayama T, Tanaka Y, Okabe T, et al. Development of a genome editing technique using the CRISPR/Cas9 system in the industrial filamentous fungus Aspergillus oryzae. Biotechnol Lett. 2016;38:637–642.
- Al Abdallah Q, Ge W, Fortwendel JR. A simple and universal system for gene manipulation in Aspergillus fumigatus: in vitro-assembled Cas9-guide RNA ribonucleoproteins coupled with microhomology repair templates. mSphere. 2017;2:e00446-17.
- Weber J, Valiante V, Nodvig CS, et al. Functional reconstitution of a fungal natural product gene cluster by advanced genome editing. ACS Synth Biol. 2017;6:62–68.
- Nakamura H, Katayama T, Okabe T, et al. Highly efficient gene targeting in Aspergillus oryzae industrial strains under ligD mutation introduced by genome editing: strain-specific differences in the effects of deleting EcdR, the negative regulator of sclerotia formation. J Gen Appl Microbiol. 2017;63:172–178.
- Nodvig CS, Hoof JB, Kogle ME, et al. Efficient oligo nucleotide mediated CRISPR-Cas9 gene editing in Aspergilli. Fungal Genet Biol. 2018;115:78–89.
- Matsuda Y, Bai T, Phippen CBW, et al. Novofumigatonin biosynthesis involves a non-heme iron-dependent endoperoxide isomerase for orthoester formation. Nat Commun. 2018;9:2587.
- Zheng X, Zheng P, Zhang K, et al. 5S rRNA promoter for guide RNA expression enabled highly efficient CRISPR/Cas9 genome editing in Aspergillus niger. ACS Synth Biol. 2019;8:1568–1574.
- Leynaud-Kieffer LMC, Curran SC, Kim I, et al. A new approach to Cas9-based genome editing in Aspergillus niger that is precise, efficient and selectable. PLoS One. 2019;14:e0210243.
- Kadooka C, Yamaguchi M, Okutsu K, et al. A CRISPR/Cas9-mediated gene knockout system in Aspergillus luchuensis mut. kawachii. Biosci Biotechnol Biochem. 2020;84:2179–2183.
