Abstract
Six interesting fungal strains were isolated during a survey of fungal diversity associated with freshwater; these strains were designated as CNUFC YJW2-22, CNUFC MSW11-6-2, CNUFC HRS5-3, CNUFC MSW242-6, CNUFC DMW2-2, and CNUFC CPWS-1. Based on a polyphasic approach including phylogenetic analyses of internal transcribed space (ITS), large subunit (LSU), beta-tubulin (BenA), and calmodulin (CaM) gene sequences, morphological analyses, the six strains were found to be identical to Acremonium guillematii, Cadophora novi-eboraci, Lectera nordwiniana, Mycoarthris corallina, Talaromyces siamensis, and Tetracladium globosum, respectively. To our knowledge, these are the first records of the rare Lectera, Mycoarthris, and Tetracladium genera in Korea, and the first reports of A. guillematii, C. novi-eboraci, L. nordwiniana, M. corallina, T. siamensis, and Te. globosum in a freshwater environment.
1. Introduction
Freshwater fungi are a diverse group of organisms best known for being ecologically important decomposers in stream ecosystems. The group is classified into families of various orders, belonging to the classes Ascomycetes, Basidiomycetes, Chytridiomycetes, Zygomycetes, and Glomeromycetes [Citation1–4]. Goh and Hyde [Citation1] reported more than 600 species of freshwater fungi. Further, the number of new taxa isolated from freshwater environments is rapidly increasing due to global contribution of reliable and complete sequencing data.
The genus Acremonium was classified by Link (1809) [Citation5] and includes some of the most simply structured of all filamentous anamorphic fungi [Citation6]. Acremonium is characterized by simple verticillate conidiophores; solitary, aculeate phialides; and unicellular conidia produced in mucoid heads or unconnected chains. Species of Acremonium are common in substrates such as soil and plant debris [Citation7,Citation8]; while some species cause important diseases in plants [Citation9,Citation10], others are agents of opportunistic infection in humans [Citation11,Citation12]. Acremonium comprises more than 100 species; however, only seven species have previously been reported in Korea: A. strictum, A. acutatum, A. zonatum, A. sclerotigenum, A. variecolor, A. persicinum, and A. tubakii [Citation13,Citation14].
The genus Cadophora was first described by Lagerberg and Melin in 1927 [Citation15] with C. fastigiata as a type species, classified based on the production of single-phialide conidiophores with distinct flask-shaped, hyaline collarettes [Citation15,Citation16]. Cadophora species have been isolated from plants [Citation16], soil [Citation17], and decaying wood [Citation18,Citation19]. Currently, the genus Cadophora comprises 17 species, of which two have been reported in Korea: C. malorum, and C. fastigiata [Citation20,Citation21].
The genus Lectera was described by Cannon in 2012 in order to reclassify Volutella colletotrichoides J.E. Chilton. Cannon [Citation22] transferred V. colletotrichoides to the genus Lectera from the genus Volutella based on morphological and molecular research, and renamed the species Lectera colletotrichoides. The species of this genus are commonly found in soil and plants such as wheat, chili, and kidney bean [Citation23]. Some species of this genus are known to be plant pathogens, and L. colletotrichoides is pathogenic to legumes [Citation22]. Currently, there are eight accepted species in this genus, according to the Index Fungorum (www.indexfungorum.org).
The genus Mycoarthris was established by Marvanová in 2002. This genus contains only one species, Mycoarthris corallina, belonging to the Calloriaceae family. It is also the type species of this genus and was isolated from foam in a stream in the United Kingdom [Citation24]. This species possesses chains of rod-like arthroconidia [Citation24]. There are no published reports of this genus in Korea.
The genus Talaromyces was established by Benjamin (1955) [Citation25] to classify a teleomorph of Penicillium, with Talaromyces vermiculatus (=T. flavus) as the type species. These species are characterized by cleistothecial or gymnothecial ascomata, unitunicate 8-spored asci, and unicellular ascospores with or without equatorial crests. Talaromyces species have been isolated from indoor air, dust, soil, clinical samples, plants, seeds, leaf litter, honey, pollen [Citation26,Citation27]. More than 150 Talaromyces species have been described worldwide [Citation28]. In Korea, only nine species of Talaromyces, belonging to sections Talaromyces and Trachyspermi, have been well described [Citation29–34].
The genus Tetracladium was described by De Wild in 1893 [Citation35] with Te. marchalianum as a type species, and it is common in aquatic habitats. Nineteen years later, Grove [Citation36] transferred Titaea maxilliformis and Tridentaria setigera to genus Tetracladium as Te. maxilliforme and Te. setigerum, respectively. Later, Te. apiense, Te. furcatum, Te. breve, Te. palmatum, and Te. nainitalense were added to the genus [Citation37,Citation38]. Recently, Te. ellipsoideum, Te. globosum, and Te. psychrophilum described by Wang et al. [Citation39] were isolated from soil in China. Currently, 11 valid species of Tetracladium are known according to the Index Fungorum, most of which produce distinctive, characteristically-shaped conidia [Citation40].
Freshwater fungi play an important role in decay of wood and leafy materials in aquatic ecosystem as well as in occurrences of plant and animal diseases, and some of them have potential to produce various antibiotics and enzymes [Citation41,Citation42]; however, only a limited number of research on freshwater fungi has been carried out in Korea [Citation33,Citation43–46]. In particular, few reports of aquatic hyphomycetes (also known as freshwater hyphomycetes or Ingoldian fungi) in Korea exist [Citation47].
The aim of the present study was to isolate and describe the six newly-recorded fungal taxa from freshwater niche using morphological and molecular analyses: A. guillematii, C. novi-eboraci, L. nordwiniana, M. corallina, T. siamensis, and Te. globosum.
2. Materials and methods
2.1. Isolation of fungal strains
Water samples were collected from the Wonhyo valley in Gwangju, the Jukrim reservoir in Yeosu, the Hwangnyong river in Gwangju, pond at Ming Ok Heon garden in Damyang, and pond located in the Chonnam National University Arboretum, Gwangju, Korea. These samples were transported to the laboratory in sterile 50-mL conical tubes and were stored at 4 °C until examination. Fungal strains were isolated by a serial dilution plating method. For dilution, 1 mL of each freshwater sample was mixed with 9 mL sterile distilled water, and the solution was shaken for 8 min. Then, serial dilutions were made ranging from 10−1 to 10−4. A 100 µL aliquot of each dilution was spread onto potato dextrose agar (PDA: 39 g of potato dextrose agar in 1 L of deionized water; Becton, Dickinson, and Co., Sparks, MD, USA) supplemented with the antibiotic neomycin (final concentration 50 ppm). The petri plates were incubated at 20–25 °C for 7–14 days. Pure isolates were obtained by selecting individual colonies of varying morphologies, transferring them to PDA plates, and subculturing until pure mycelia were obtained. For stock storage, the pure isolates were maintained in PDA slant tubes and 20% glycerol at −80 °C at the Environmental Microbiology Laboratory Fungarium, Chonnam National University, Gwangju, Korea. The strains CNUFC MSW11-6-2, CNUFC MSW242-6, CNUFC YJW2-22, CNUFC HRS5-3, CNUFC DMW2-2, and CNUFC CPWS-1 were also deposited at the Culture Collection of the Nakdonggang National Institute of Biological Resources (NNIBR, Sangju, Korea).
2.2. Morphological examination
To observe morphology, the CNUFC MSW11-6-2, CNUFC MSW242-6, CNUFC YJW2-22, and CNUFC HRS5-3 strains were cultured on PDA, malt extract agar (MEA; malt extract 20 g, agar 20 g in 1 L of deionized water), and oatmeal agar (OA; oat meal 30 g, agar 15 g in 1 L of deionized water). CNUFC DMW2-2 was culture on czapek yeast autolysate agar (CYA), yeast extract sucrose agar (YES), and Blakeslee’s malt extract agar [Citation48]. CNUFC CPWS-1 was culture on PDA. All plates were incubated at 25 °C in the dark for 13–14 days, except for CNUFC CPWS1 was incubated at 20 °C. Samples were mounted in lactophenol solution (Junsei Chemical Co. Ltd., Tokyo, Japan) or distilled water before being observed under an Olympus BX51 and BX53 microscope with DIC optics (Olympus, Tokyo, Japan).
2.3. DNA extraction, PCR, and sequencing
Genomic DNA was extracted directly from the mycelia and spores of fungal isolates using the Solg Genomic DNA Prep Kit (Solgent Co., Ltd., Daejeon, South Korea). The internal transcribed spacer (ITS) region, large subunit (LSU), beta-tubulin (BenA), and calmodulin (CaM) genes were amplified using the primer pairs ITS5 (5′-GGAAGTAAAAGTCGTAACAAGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) [Citation49], LROR (5′-ACCCGCTGAACTTAAGC-3′) and LR5F (5′-GCTATCCTGAGGGAAAC-3′) [Citation50], Bt2a (5′-GGTAACCAAATCGGTGCTGCTTTC-3′) and Bt2b (5′-ACCCTCAGTGTAGTGACCCTTGGC-3′) [Citation51], and CF1 (5′-GCCGACTCTTTGACY GARGAR-3′) and CF4 (5′-TTTYTGCATCATRAGYTGGAC-3′) [Citation52], respectively. The PCR products were purified using the Accuprep PCR Purification Kit (Bioneer Corp.). Sequencing was performed using the same PCR primers and was run on the ABI PRISM 3730XL Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
2.4. Phylogenetic analysis
Fungal strain sequences were initially aligned using Clustal X v.2.0 [Citation53], and the alignment was edited using BioEdit v.7.2.6 [Citation54]. Phylogenetic analyses were conducted using MEGA 7 [Citation55], and phylogenetic trees were made based on the maximum likelihood (ML) method and the Kimura 2-parameter model. The reliability of internal branches was assessed using the p-distance substitution model with 1,000 bootstrap replications. The sequences of CNUFC MSW11-6-2, CNUFC MSW11-6-2-1, CNUFC MSW242-6, CNUFC MSW242-8, CNUFC YJW2-22, CNUFC YJW2-22-1, CNUFC HRS5-3, CNUFC HRS5-3-1, CNUFC DMW2-2, CNUFC DMW2-2-1, CNUFC CPWS-1, and CNUFC CPWS-2 were deposited in the NCBI database under the accession numbers shown in .
Table 1. Sequences used in this study and GenBank accession numbers.
3. Results
3.1. Phylogenetic analysis
The ITS sequence analysis by the Basic Local Alignment Search Tool (BLAST) indicated that the CNUFC MSW11-6-2, CNUFC MSW242-6, CNUFC YJW2-22, CNUFC HRS5-3, CNUFC DMW2-2, and CNUFC CPWS-1 isolates were 99.7% (584/586 bp), 99.8% (416/417 bp), 98.8% (513/519 bp), 99.1% (520/525 bp), 99.4% (516/519 bp), and 100% (417/417 bp) related to C. novi-eboraci (GenBank accession no. KM497036), M. corallina (GenBank accession no. AH009124), A. guillematii (GenBank accession no. AB828055), L. nordwiniana (GenBank accession no. MK047462), T. siamensis A3S2-40 (GenBank accession no. KJ767055), and Te. globosum MY25 (GenBank accession no. JX029133), respectively. The 28S sequence of CNUFC YJW2-22 and CNUFC HRS5-3 showed similarities of 99.4% (774/779 bp), and 99.7% (846/849 bp) with A. guillematii CBS 767.69 (GenBank accession no. MH871190), and L. nordwiniana CBS 144922 (GenBank accession no. MK047513). BLASTn analysis of BenA of CNUFC DMW2-2 and CNUFC MSW11-6-2 showed similarities of 100% (399/399 bp), and 99.5% (607/610 bp) with T. siamensis CBS 475.88 (GenBank accession no. JX091379), and C. novi-eboraci GLMC 274 (GenBank accession no. MN232975). Similarity, BLASTn analysis of CaM of CNUFC DMW2-2 showed similarities of 99.6% (470/472 bp) with T. siamensis DTO 269I3 (GenBank accession no. KJ775428). Based on the ITS, LSU, BenA, and a combined (ITS + BenA + CaM) trees, CNUFC YJW2-22, CNUFC MSW11-6-2, CNUFC HRS5-3, CNUFC MSW242-6, CNUFC DMW2-2, and CNUFC CPWS-1 isolates were identical to A. guillematii, C. novi-eboraci, L. nordwiniana, M. corallina, T. siamens, and Te. globosum, respectively ().
Figure 1. Phylogenetic tree of Acremonium guillematii CNUFC YJW2-22 and CNUFC YJW2-22-1, and related species, based on a maximum likelihood analyses of internal transcribed spacer (A) and large subunit (B) sequences. The sequence of Plectosphaerella cucumerina was used as an outgroup. Bootstrap support values of ≥50% are indicated at the nodes. Ex-type strains are indicated by T.
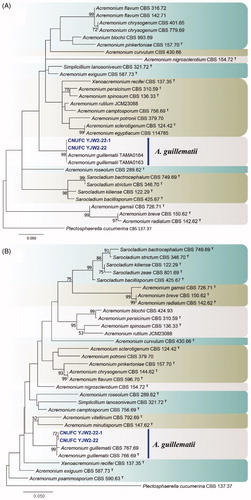
Figure 2. Phylogenetic tree of Cadophora novi-eboraci CNUFC MSW11-6-2 and CNUFC MSW11-6-2-1, and related species, based on maximum likelihood analyses of internal transcribed spacer (A) and beta-tubulin (B) sequences. The sequence of Cadophora finlandica was used as an outgroup. Bootstrap support values of ≥50% are indicated at the nodes. Ex-type strains are indicated by T.
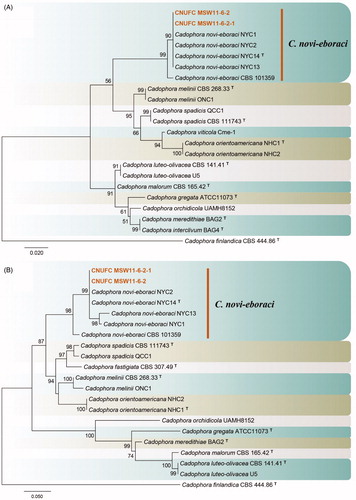
Figure 3. Phylogenetic tree of Lectera nordwiniana CNUFC HRS5-3 and CNUFC HRS5-3-1, and related species, based on maximum-likelihood analyses of internal transcribed spacer (A) and large subunit (B) sequences. The sequence of Monilochaetes melastomae was used as an outgroup. Bootstrap support values of ≥50% are indicated at the nodes. Ex-type and ex-neotype strains are indicated by T and NT.
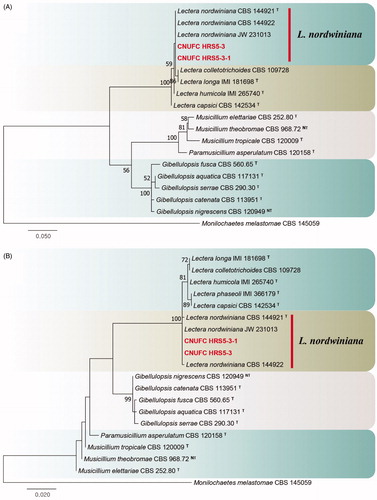
Figure 4. Phylogenetic tree of Mycoarthris corallina CNUFC MSW242-6 and CNUFC MSW242-8, and related species, based on maximum-likelihood analysis of internal transcribed spacer sequences. The sequences of Monilinia laxa and Sclerotinia sclerotiorum were used as outgroups. Bootstrap support values of ≥50% are indicated at the nodes. Ex-type strains are indicated by T.
Figure 5. Phylogenetic tree of Talaromyces siamensis CNUFC DMW2-2 and CNUFC DMW2-2-1, and related species, based on maximum-likelihood analyses of internal transcribed spacer, beta-tubulin, and calmodulin sequences. The sequence of Talaromyces dendriticus was used as an outgroup. Bootstrap support values of ≥50% are indicated at the nodes. Ex-type strains are indicated by T.
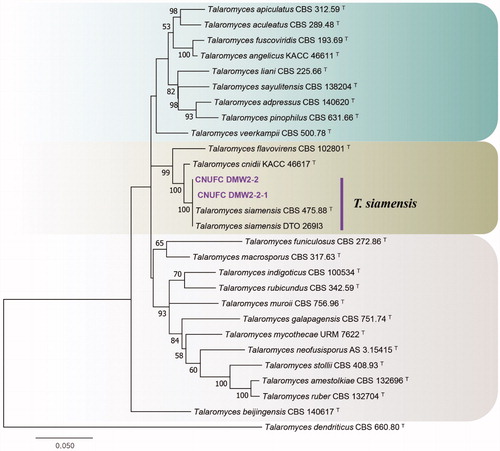
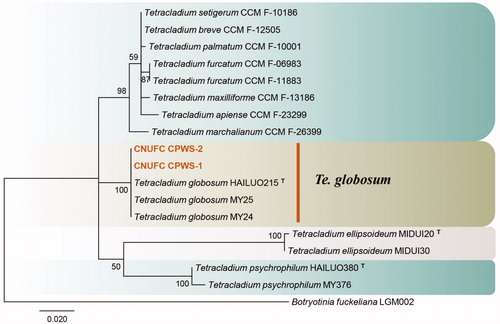
Figure 6. Phylogenetic tree of Tetracladium globosum CNUFC CPWS-1 and CNUFC CPWS-2, and related species, based on maximum-likelihood analysis of internal transcribed spacer sequences. The sequence of Botryotinia fuckeliana was used as an outgroup. Bootstrap support values of ≥50% are indicated at the nodes. Ex-type strains are indicated by T.
3.2. Taxonomy
3.2.1. Taxonomy of CNUFC YJW2-22
Acremonium guillematii Gams, Cephalosporium-artige Schimmelpilze: 56 (1971) [Citation56] ().
Figure 7. Morphology of Acremonium guillematii CNUFC YJW2-22. (A,D) Colonies on potato dextrose agar. (B,E) Colonies on malt extract agar. (C,F) Colonies on oatmeal agar. (A–C) obverse view, (D–F) reverse view. (G,H) Branched conidiophores. (I,J) Conidiophores and clustered conidia in slimy heads. (K) Octahedral crystals in culture. (L) Conidia. Scale bars: G-K = 20 µm, L = 10 µm.
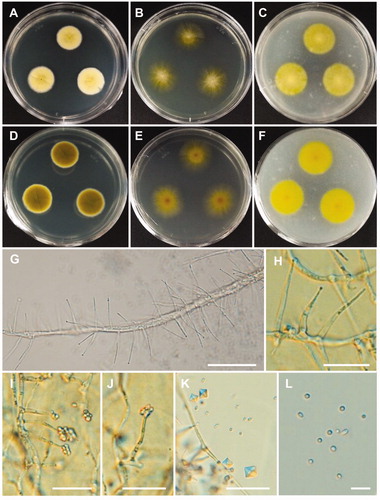
Description: Colonies grew slowly on 2% MEA in the dark at 25 °C, reaching 23–26 mm in diameter over 14 days and were ivory to intense yellow with a flat, velvety texture. The reverse side of the colonies was intense yellow. Conidophores were solitary, sparsely branched, and orthotropic with slender phialides, and measured 11.5–31.5 × 1.3–2.7 µm. Conidia were globose, subglobose, ellipsoidal or ovoid, and measured 1.6–3.7 × 1.5–2.5 µm.
3.2.2. Taxonomy of CNUFC MSW11-6-2
Cadophora novi-eboraci Travadon, Lawr, Roon-Lath, Gubler, Wilcox, Rolsh & Baumgartner, Fungal Biology 119: 61 (2015) [Citation16] ().
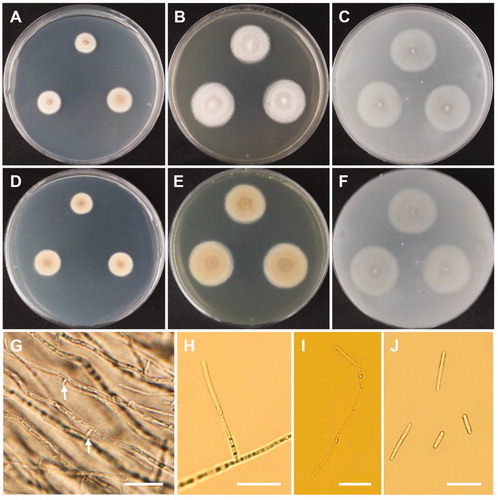
Figure 8. Morphology of Cadophora novi-eboraci CNUFC MSW11-6-2. (A,D) Colonies on potato dextrose agar. (B,E) Colonies on malt extract agar. (C,F) Colonies on oatmeal agar. (A–C) obverse view, (D–F) reverse view. (G–I) Conidiogenous cells and conidia in slimy heads. (J) Conidia. Scale bars: G-J = 10 µm.
Description: Colonies grew slowly on 2% MEA in the dark at 25 °C, reaching 13–14 mm in diameter over 13 days, and appeared deep orange with a flat texture and smooth margins. The reverse side of each colony was deep orange. Conidiophores were simple or branched, hyaline, and measured 8.3–69 × 1.5–3 µm. Conidiogenous cells were terminal or lateral, commonly solitary, obclavate, ampulliform, subcylindrical, flexuous, and measured 4–15 × 1–3 µm. Collarettes were short, flaring, cup–shaped, parallel, and measured 1.5–2 × 1–1.8 µm. Conidia were ovoid, cylindrical to oblong, elliptical, and measured 3–6 × 1–4 µm.
3.2.3. Taxonomy of CNUFC HRS5-3
Lectera nordwiniana Crous, Luangsa-Ard, Wingfield, et al., Persoonia 41: 349 (2018) [Citation57] ().
Figure 9. Morphology of Lectera nordwiniana CNUFC HRS5-3. (A,D) Colonies on potato dextrose agar. (B,E) Colonies on malt extract agar. (C,F) Colonies on oatmeal agar. (A–C) obverse view, (D–F) reverse view. (G) Young conidiomata on potato dextrose agar. (H,I) Conidiomata with setae. (J) Apex of a seta. (K,L) Phialides with conidia on acervuli. (M) Conidia. Scale bars: G = 1 mm, H = 200 µm, I = 20 µm, J-M = 10 µm.
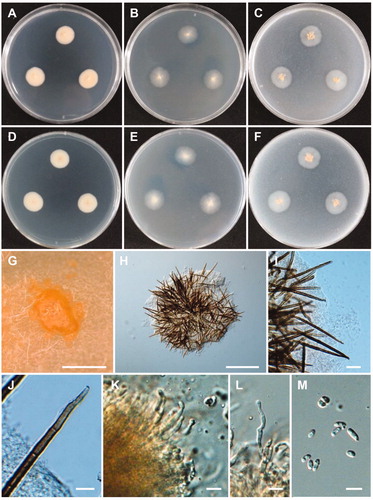
Description: Colonies grew slowly on PDA in the dark at 25 °C, reaching 21–23 mm in diameter over 14 days, were beige to bright salmon and were slightly elevated. The reverse sides of the colonies were beige to bright salmon in color. Conidiomata were salmon in color, sporodochial, punctiform, solitary or gregarious and were abundantly formed on PDA, 2% MEA, and OA. Setae were dark brown, 3–8 septate, flexuous or rigid, tapering to acutely rounded apices, surrounding conidiomata, and measured 57–205 × 4–6.4 µm. Phialides were subcylindrical to subulate, hyaline, smooth–walled, and measured 12.5–29 × 1.8–3.5 µm. Conidia were fusiform to navicular, hyaline, orange in mass, and measured 4.5–9 × 2–4.5 µm.
3.2.4. Taxonomy of CNUFC MSW242-6
Mycoarthris corallina Marvanová & Fisher, Nova Hedwigia 75: 258 (2002) [Citation24] ().
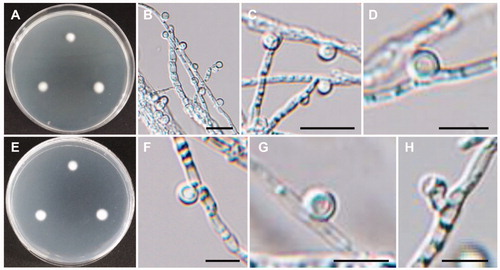
Figure 10. Morphology of Mycoarthris corallina CNUFC MSW242-6. (A,D) Colonies on potato dextrose agar. (B,E) Colonies on malt extract agar. (C,F) Colonies on oatmeal agar. (A–C) obverse view, (D–F) reverse view. (G) Anastomosing hyphae (white arrows). (H) Conidiophore. (I) Chain of arthroconidia. (J) Conidia. Scale bars: G-J = 20 µm.
Description: Colonies grew slowly on 2% MEA in the dark at 15 °C, reaching 18 mm in diameter over 13 days, were off–white in color, and were flat. The reverse side of each colony was pale beige. Conidiophores were terminal or lateral, simple or sparsely branched, hyaline, and measured up to 39 × 2.5–3 µm. Conidia were cylindrical or slightly curved, produced a chain of arthroconidia, and measured 9.5–36 × 1.5–2.5 µm.
3.2.5. Taxonomy of CNUFC DMW2-2
Talaromyces siamensis (Manoch & Ramírez) Samson, Yilmaz & Frisvad, Studies in Mycology 70: 177 (2011) [Citation26] ().
Figure 11. Morphology of Talaromyces siamensis CNUFC DMW2-2. (A,D) Colonies on czapek yeast autolysate agar. (B,E) Colonies on yeast extract sucrose agar. (C,F) Colonies on malt extract agar. (A–C) obverse view, (D–F) reverse view. (G–J) Conidiophores. (K) Conidia. Scale bars: G-J = 20 µm, K = 10 µm.
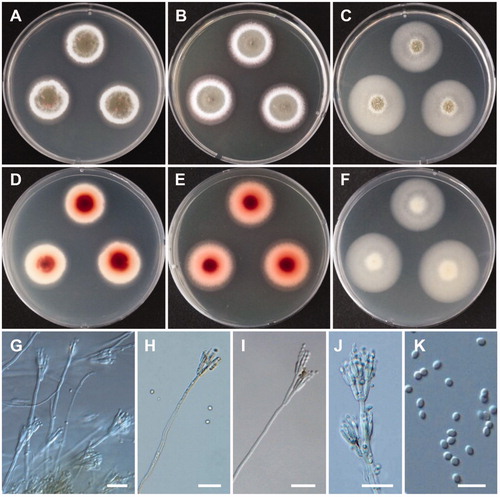
Description: Colonies grew slowly on CYA in the dark at 25 °C, reaching 30 mm in diameter over 7 days and were olive green. The reverse side of each colony was reddish at the center and faded into a bright red. Conidiophores were biverticillate, solitary or branched, with smooth walled stipes, and measured 115–475 × 2.8–4 µm. Metulae were three to six divergent and measured 9.3–16 × 2.2–3.5 µm. Phialides were acerose, numbered three to five per metulae, and measured 9–14.5 × 2–3 µm. Conidia were ovoid, ellipsoidal to fusiform, and measured 2.8–4 × 2–3 µm.
3.2.6. Taxonomy of CNUFC CPWS-1
Tetracladium globosum M.M. Wang & Xing Z. Liu, Persoonia 34: 108 (2015) [Citation39] ().
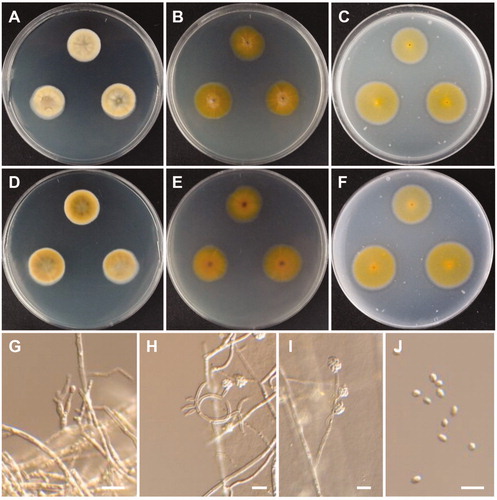
Figure 12. Morphology of Tetracladium globosum CNUFC CPWS-1. (A,E) Colonies on potato dextrose agar. (A) obverse view, (E) reverse view. (B–D) Conidia and hyphae. (F–H) Conidia. Scale bars: B-C = 20 µm, D, F-H = 10 µm.
Description: Colonies grew slowly on PDA, reaching 8 mm in diameter over 7 days at 15 °C. The colony color was pale yellow. Vegetative hyphae possessed transverse septa, measured 1–4 µm, and were smooth, thin walled, and hyaline. Conidia were globose, smooth-walled, hyaline, measured 4.7–6.5 µm, and were attached to the hyphae with short conidiophores.
4. Discussion
To date, few studies have reported undescribed species in Korean freshwater environments. Therefore, collecting freshwater samples to identify undescribed fungal species is the best way to enhance our knowledge of Korean filamentous fungi. The discovery of A. guillematii, C. novi-eboraci, L. nordwiniana, M. corallina, T. siamensis, and Te. globosum in the present study improves our knowledge of the occurrence and distribution of freshwater fungi in Korea, especially, rare genera of Lectera, Mycoarthris, and Tetracladium.
The ITS rDNA region, which is regarded as barcode marker, is well-established as a method for identifying fungal taxa [Citation58]. Additionally, LSU and β-tubulin have previously been used to accurately identify fungal strains at the species level. In the present study, the CNUFC YJW2-22 and CNUFC YJW2-22-1 isolates were identified to A. guillematii based on ITS and LSU sequence data (). The morphological features of the CNUFC YJW2-22 isolate were similar to those of A. guillematii, as described by Gams [Citation56]. In the current study, the strain produced yellow colonies and octahedral crystals on 2% MEA, which was found to be similar to A. guillematii [Citation56,Citation59]. Many Acremonium species have been identified as useful fungal resources as they produce bioactive metabolites. Tian et al. [Citation60] reviewed the secondary metabolites of Acremonium species and revealed a total of 356 metabolites including steroids, terpenoids, polyketides, meroterpenoids, peptides, and alkaloids.
The analyses of ITS and BenA gene sequences showed that the CNUFC MSW11-6-2 and CNUFC-MSW11-6-2-1 isolates were clustered with C. novi-eboraci (type strain), with well-supported branches, in full agreement with the results of Travadon et al. [Citation16] (). The CNUFC MSW11-6-2 isolate was most morphologically-similar to C. novi-eboraci, as described by Travadon et al. [Citation16]. Cadophora luteo-olivacea isolated from huts in Antarctica have been reported to produce cadopherone and colomitide polyketides [Citation61]. In addition, Almeida et al. [Citation62] isolated four types of hydroxylated sclerosporin from the marine-derived Cadophora malorum fungus.
Phylogenetic analysis of ITS and LSU sequences showed that the CNUFC HRS5-3 and CNUFC HRS5-3-1 strains were clustered within the same clade as L. nordwiniana (type strain) (). Molecular data analysis results were consistent with the phylogeny presented by Giraldo and Crous [Citation63] and Crous et al. [Citation57]. The morphological characteristics of the CNUFC HRS5-3 isolate were generally similar to those previously described by Crous et al. [Citation57] such as long flexuous setae (up to 205 µm). However, the observations of the current study showed that the CNUFC HRS5-3 isolate produced salmon conidiomata, whereas L. nordwiniana (type strain) produces brown conidiomata [Citation57] Interestingly, the strain isolated in the present produced extracellular enzymes including protease and lipase (data not shown). Therefore, the Lectera species obtained in this study may have useful applications based on this property. L. nordwiniana was previously isolated from Dutch soils [Citation57], making the present study the first instance of the species being recovered from freshwater.
Phylogenetic analyses revealed that the CNUFC MSW242-6 and CNUFC MSW242-8 isolates were clustered with M. corallina (type strain) (). The CNUFC MSW242-6 isolate was morphologically similar to M. corallina as it produced arthroconidia on 2% MEA medium [Citation24]. Similar to M. corallina isolated by Marvanová et al. [Citation24], the strain was isolated from stream foam in the present study. Therefore, stream foam is a habitat for M. corallina.
The phylogenetic tree created using ITS, BenA, and CaM sequences showed that the CNUFC DMWW2-2 and CNUFC DMWW2-2-1 isolates were placed within the T. siamensis clade, belonging to the section Talaromyces (). The morphological characteristics of the T. siamensis isolate in this study were similar to those previously described by Yilma et al. [Citation27]. However, differences in colony diameter were observed on CYA (CYA: 20–22 mm after 7 days) and MEA (MEA: 32–33 mm after 7 days) media. T. siamensis is known to produce mitorubrins, penicillides, purpactins, vermixocins, secalonic acids D & F, and vermicellin [Citation27], and the species was previously isolated from forest soil and house dust from Thailand [Citation27]. The present study describes the first instance of T. siamensis isolated from freshwater.
Molecular phylogenetic analysis using ITS sequence indicated that CNUFC CPWS-1 and CNUFC CPWS-2 were grouped with Te. globosum (type strain) (), and the molecular data analyses of these species were consistent with the phylogeny presented by Wang et al. [Citation39]. The CNUFC CPWS-1 isolate is morphologically-similar to Te. globosum as it produces globose and 1-celled conidia. However, the conidia sizes reported in the literature (3.0–5.5 µm) are slightly smaller than those of the isolates in the present study. Te. globosom was first isolated in a glacial soil sample from the Hailuogou Glacier in China, one of the most extreme environments on earth [Citation39]. Te. globosum species has been isolated for the first time from freshwater in Korea. The occurrence of the species in such freshwater niche suggests that it plays an important role in aquatic ecosystems.
Recently, novel species in Penicillium (Penicillium acidum Hyang B. Lee, T.T. Duong & T.T.T. Nguyen, and Penicillium aquaticum Hyang B. Lee, T.T. Duong & T.T.T. Nguyen), Rhizophydium (Rhizophydium koreanum Hyang B. Lee, S.J. Jeon, T.T.T. Nguyen), and Mucor (Mucor fluvii Hyang B. Lee, S.H. Lee & T.T.T. Nguyen) have been discovered from freshwater niche in Korea [Citation64,Citation65]. A number of undescribed fungal species were also reported from freshwater [Citation33,Citation43–46]. However, only a small number of aquatic hyphomycetes were found in freshwater [Citation47]. Therefore, a large number of undescribed fungal taxa in freshwater still remain undiscovered on planet. Future research focusing on aquatic ascomycetes or aquatic hyphomycetes known as Ingoldian fungi in Korea are needed to improve our understanding of the ecology of these groups, especially rare genera.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Goh TK, Hyde K. Biodiversity of freshwater fungi. J in Microbiol Biot. 1996;17(5-6):328–345.
- Shearer CA. The freshwater Ascomycetes. Nova Hedwigia. 1993;56:1–33.
- Shearer CA, Descals E, Kohlmeyer B, et al. Fungal biodiversity in aquatic habitats. Biodivers Conserv. 2007;16(1):49–67.
- Jones EBG, Hyde KD, Pang KL. Freshwater fungi and fungal-like organisms. Boston: De Gruyter; 2014.
- Link HF. Observationes in ordines plantarum naturales. Dissertatio I. Magazin der Gesellschaft Naturforschenden Freunde Berlin, 1809.
- Summerbell RC, Gueidan C, Schroers HJ, et al. Acremonium phylogenetic overview and revision of Gliomastix, Sarocladium, and Trichothecium. Stud Mycol. 2011;68:139–162.
- Domsch KH, Gams W, Anderson TH. Compendium of soil fungi. Eching: IHW-Verlag; 2007. p. 30–38.
- Watanabe T, Watanabe Y, Fukatsu T. New species of Acremonium, Cylindrocarpon and Verticillium from soil in the Bonin (Ogasawara) Islands, Japan. Mycoscience. 2001;42(6):591–595.
- Alfaro-García A, Armengol J, Bruton BD, et al. The taxonomic position of the causal agent of Acremonium collapse of muskmelon. Mycologia. 1996;88(5):804–808.
- Lin HJ, Chien CY, Huang JW. Pathogenicity and host range of Acremonium lactucae sp. nov., the causal agent of leaf brown spot of lettuce. Plant Pathol Bull. 2004;13:91–96.
- Summerbell RC. Ascomycetes: Aspergillus, Fusarium, Sporothrix, Piedraia, and their relatives. In: Howard DH, editor. Pathogenic fungi in humans and animals. 2nd ed. New York: Marcel Dekker; 2003. p. 237–498.
- de Hoog GS, Guarro J, Gené J, et al. Atlas of clinical fungi. USB version 4.1. Utrecht: CBS-KNAW Fungal Biodiversity Centre; 2015.
- Park SW, Nguyen TTT, Lee HB. Characterization of two species of Acremonium (unrecorded in Korea) from soil samples: A. variecolor and A. persicinum. Mycobiology. 2017;45(4):353–361.
- Oh Y, Mun HY, Goh J, et al. New records of three Sordariomycetes fungi isolated from aquatic plants in Jeju, Korea. Kor J Mycol. 2018;46:91–97.
- Lagerberg T, Melin E, Lundberg G. Biological and practical researches into blueing in pine and spruce. Svenska Skogsvdrdsforen Tidskr. 1927;25:145–272.
- Travadon R, Lawrence DP, Rooney-Latham S, et al. Cadophora species associated with wood-decay of grapevine in North America. Fungal Biol. 2015;119(1):53–66.
- Agustí-Brisach C, Gramaje D, García-Jiménez J, et al. Detection of black-foot and petri disease pathogens in soils of grapevine nurseries and vineyards using bait plants. Plant Soil. 2013;364(1-2):5–18.
- Blanchette RA, Held BW, Jurgens JA, et al. Wood-destroying soft rot fungi in the historic expedition huts of Antarctica. Appl Environ Microbiol. 2004;70(3):1328–1335.
- Blanchette RA, Held BW, Arenz BE, et al. An Antarctic hot spot for fungi at Shackleton's historic hut on cape royds. Microb Ecol. 2010;60(1):29–38.
- You YH, Yoon H, Kang SM, et al. Cadophora malorum Cs-8-1 as a new fungal strain producing gibberellins isolated from Calystegia soldanella. J Basic Microbiol. 2013;53(7):630–634.
- Park KM, Ku KH, Koo M. Antifungal activity of plant extracts against Fusarium oxysporum isolated from spoilage ginger. Kor J Food Preserv. 2018;25(2):173–180.
- Cannon PF, Buddie AG, Bridge PD, et al. Lectera, a new genus of the Plectosphaerellaceae for the legume pathogen Volutella colletotrichoides. MycoKeys. 2012;3:23–36.
- Crous PW, Wingfield MJ, Burgess TI, et al. Fungal Planet description sheets: 558-624. Persoonia. 2017;38:240–384.
- Marvanová L, Landvik S, Fisher PJ, et al. A new fungus with arthroconidia from foam. Nova Hedw. 2002;75(1):255–269.
- Benjamin CR. Ascocarps of Aspergillus and Penicillium. Mycologia. 1955;47(5):669–687.
- Samson RA, Yilmaz N, Houbraken J, et al. Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Stud Mycol. 2011;70(1):159–183.
- Yilmaz N, Visagie CM, Houbraken J, et al. Polyphasic taxonomy of the genus Talaromyces. Stud Mycol. 2014; 78:175–341.
- Sun BD, Chen AJ, Houbraken J, et al. New section and species in Talaromyces. MycoKeys. 2020;768:75–113.
- Sang H, An T, Kim CS, et al. Two novel Talaromyces species isolated from medicinal crops in Korea. J Microbiol. 2013;51(5):704–708.
- Adhikari M, Yadav DR, Kim S, et al. Discovery of two new Talaromyces species from crop field soil in Korea. Mycobiology. 2015;43(4):402–407.
- Nguyen TTT, Paul NC, Lee HB. Characterization of Paecilomyces variotii and Talaromyces amestolkiae in Korea based on the morphological characteristics and multigene phylogenetic analyses. Mycobiology. 2016;44(4):248–259.
- Pangging M, Nguyen TTT, Lee HB. New records of four species belonging to Eurotiales from soil and freshwater in Korea. Mycobiology. 2019;47(2):154–164.
- Heo I, Hong K, Yang H, et al. Diversity of Aspergillus, Penicillium, and Talaromyces species isolated from freshwater environments in Korea. Mycobiology. 2019;47(1):12–19.
- You YH, Aktaruzzaman M, Heo I, et al. Talaromyces halophytorum sp. nov. isolated from roots of Limonium tetragonum in Korea. Mycobiology. 2020;48(2):133–138.
- Wildeman E. d. Notes Mycologiques. Fasc. 2. Annales de la Societé Belge de Microscopie. 1893;17:35–68.
- Grove WB. New or noteworthy fungi, Part IV. J Bot. 1912;50:9–18.
- Descals E, Webster J. Four new staurosporous hyphomycetes from mountain streams. Trans Brit Mycol Soc. 1983;69:80–109.
- Sati SC, Arya P, Belwal M. Tetracladium nainitalense sp. nov, a root endophyte from Kumaun Himalaya, India. Mycologia. 2009;101(5):692–695.
- Wang M, Jiang X, Wu W, et al. Psychrophilic fungi from the world's roof. Persoonia. 2015;34:100–112.
- Letourneau A, Seena S, Marvanová L, et al. Potential use of barcoding to identify aquatic hyphomycetes. Fungal Divers. 2010;40(1):51–64.
- Harrigan GG, Armentrout BL, Gloer JB, et al. Anguillosporal, a new antibacterial and antifungal metabolite from the freshwater fungus Anguillospora longissima. J Nat Prod. 1995;58(9):1467–1469.
- Ho WH, To PC, Hyde KD. Induction of antibiotic production of freshwater fungi using mix-culture fermentation. Fungal Divers. 2003;12:45–51.
- Nguyen TTT, Lee SH, Jeon SJ, et al. First records of rare Ascomycete fungi, Acrostalagmus luteoalbus, Bartalinia robillardoides, and Collariella carteri from freshwater samples in Korea. Mycobiology. 2019;47(1):1–11.
- Goh J, Mun HY, Jeon YJ, et al. First report of six Sordariomycetes fungi isolated from plant litter in freshwater ecosystems of Korea. Kor J Mycol. 2020;48(2):103–116.
- Nam B, Lee JS, Lee HB, et al. Pezizomycotina (Ascomycota) fungi isolated from freshwater environments of Korea: Cladorrhinum australe, Curvularia muehlenbeckiae, Curvularia pseudobrachyspora, and Diaporthe longicolla. Kor J Mycol. 2020;48(1):29–38.
- Nguyen TTT, Jeon YJ, Mun HY, et al. Isolation and characterization of four unrecorded Mucor species in Korea. Mycobiology. 2020;48(1):29–36.
- Mun HY, Goh J, Yoosun O, et al. New records of three aquatic fungi isolated from freshwater in Samcheok and Yeongju, Korea. Kor J Mycol. 2016; 44:247–251.
- Samson RA, Houbraken J, Thrane U, et al. Food and indoor fungi. CBS laboratory manual series 2. Utrecht: CBS KNAW Fungal Biodiversity Centre. 2010. p. 390.
- White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York: Academic Press; 1990. p. 315–322.
- Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172(8):4238–4246.
- Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 1995;61(4):1323–1330.
- Peterson SW, Vega FE, Posada F, et al. Penicillium coffeae, a new endophytic species isolated from a coffee plant and its phylogenetic relationship to P. fellutanum, P. thiersii and P. brocae based on parsimony analysis of multilocus DNA sequences. Mycologia. 2005;97(3):659–666.
- Thompson JD, Gibson TJ, Plewniak F, et al. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882.
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98.
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874.
- Gams W. Cephalosporium-artige Schimmelpilze (Hyphomycetes). Verlag Stuttgart: Gustav Fischer; 1971. p. 262.
- Crous PW, Luangsa-Ard JJ, Wingfield MJ, et al. Fungal planet description sheets: 785-867. Persoonia. 2018;41:238–417.
- Schoch CL, Seifert KA, Huhndorf S, et al.; Fungal Barcoding Consortium Author List. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci USA. 2012;109(16):6241–6246.
- Yabuki T, Duncan I, Okuda T. Comparative study reveals unique features of the mycobiota in peat soils samples from Japan and Scotland. Mycoscience. 2014;55(3):168–176.
- Tian J, Lai D, Zhou L. Secondary metabolites from Acremonium fungi: Diverse structures and bioactivities. Mini Rev Med Chem. 2017;17(7):603–632.
- Rusman Y, Held BW, Blanchette RA, et al. Cadopherone and colomitide polyketides from Cadophora wood-rot fungi associated with historic expedition huts in Antarctica. Phytochemistry. 2018;148:1–10.
- Almeida C, Eguereva E, Kehraus S, et al. Hydroxylated sclerosporin derivatives from the marine-derived fungus Cadophora malorum. J Nat Prod. 2010;73(3):476–478.
- Giraldo A, Crous PW. Inside plectosphaerellaceae. Stud Mycol. 2019;92:227–286.
- Hyde KD, Dong Y, Phookamsak R, et al. Fungal diversity notes 1151–1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2020;100(1):5–277.
- Wanasinghe DN, Phukhamsakda C, Hyde KD, et al. Fungal diversity notes 709–839: taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers. 2018;89(1):1–236.
