Abstract
Despite recent studies, relatively few are known about the diversity of fungal communities in the deep Atlantic Ocean. In this study, we investigated the diversity of fungal communities in 15 different deep-sea sediments from the South Atlantic Ocean with a culture-dependent approach followed by phylogenetic analysis of ITS sequences. A total of 29 fungal strains were isolated from the 15 deep-sea sediments. These strains belong to four fungal genera, including Aspergillus, Cladosporium, Penicillium, and Alternaria. Penicillium, accounting for 44.8% of the total fungal isolates, was a dominant genus. The antiaflatoxigenic activity of these deep-sea fungal isolates was studied. Surprisingly, most of the strains showed moderate to strong antiaflatoxigenic activity. Four isolates, belonging to species of Penicillium polonicum, Penicillium chrysogenum, Aspergillus versicolor, and Cladosporium cladosporioides, could completely inhibit not only the mycelial growth of Aspergillus parasiticus mutant strain NFRI-95, but also the aflatoxin production. To our knowledge, this is the first report to investigate the antiaflatoxigenic activity of culturable deep-sea fungi. Our results provide new insights into the community composition of fungi in the deep South Atlantic Ocean. The high proportion of strains that displayed antiaflatoxigenic activity demonstrates that deep-sea fungi from the Atlantic Ocean are valuable resources for mining bioactive compounds.
1. Introduction
Marine fungi are microorganisms widely distributed in the ocean and are particularly associated with sediment, seawater, marine habitants, submerged plants, and algae [Citation1]. They are a rich source of natural products [Citation2–4]. The deep sea is one of the least explored regions of the earth and one of the least studied habitats of fungi. It is an environment characterized by the absence of sunlight irradiation, low temperature (except for hydrothermal vents), high hydrostatic pressure, and extreme pH [Citation5]. The extreme environment has created a variety of unique organisms. However, compared to the increasing knowledge about the biodiversity and ecological importance of deep-sea bacteria and archaea, relatively little is now known about deep-sea fungi.
Since the first report of the isolation of deep-sea fungi from the South Atlantic Ocean at a depth of 4450 m [Citation6], there have been many more studies noting the isolation of deep-sea fungi from various deep-sea environments and their diversity [Citation7–15]. It is now well known that diverse fungal communities are abundant in deep-sea environments. Fungi have been found in different deep-sea samples, such as sediments from the Mariana Trench [Citation8,Citation16], Chagos Trench [Citation17], the Central Indian Basin [Citation18,Citation19], the South China Sea [Citation10,Citation14], Peru Trench [Citation9], Gulf of Mexico [Citation20], Pacific Ocean [Citation21,Citation22], and the Antarctica Ocean [Citation23]. Fungal diversity in deep-sea sediments has been analyzed by either culture-dependent [Citation24] or culture-independent [Citation19,Citation21] methods. Using culture-independent molecular techniques, culturable and unculturable fungi in the environmental DNA samples could both be directly detected, thus acquiring abundant information about their biodiversity. In contrast, the culture-dependent approach is only applicable to cultivable fungi, which may underestimate the real diversity of fungal communities; however, the isolated fungi could be subjected to physiological and biochemical tests and bioactivity screening.
The potent mycotoxin aflatoxin B1 is a secondary metabolite mainly produced by Aspergillus flavus and Aspergillus parasiticus [Citation25]. It is extremely toxic, carcinogenic, and mutagenic to both humans and animals. Aflatoxin contamination can occur during the transport and storage of food products, and even before the harvest of crops. To prevent and manage aflatoxin contamination, approaches such as chemical fungicides and biological control are suggested. Biocontrol agents could be good alternative to chemical fungicides, as they are beneficial to both people and the environment. During the last decade, some bacteria and fungi have been reported for their ability in the control of aflatoxin producing fungi. For example, Bacillus pumilus, isolated from Korean soybean sauce, was reported to exhibit strong antifungal activity against the aflatoxin-producing fungi As. flavus and As. parasiticus [Citation26]. B. megaterium, an offshore bacterium isolated from the Yellow Sea of East China, was reported to be able to inhibit the growth of As. flavus in vitro and in vivo [Citation27]. Recently, our group also reported that a deep-sea bacterium B. circulans could inhibit both the mycelial growth of As. parasiticus mutant strain NFRI-95 and accumulation of norsolorinic acid, a precursor for aflatoxin production [Citation28].
Since the first report on the metabolites of the deep-sea fungus Chromocleista sp. [Citation29], more than 200 new biologically active secondary metabolites of deep-sea fungi have been isolated [Citation30]. The bioactive metabolites are structurally diverse, and some of them exhibit potential bioactivities against pathogenic fungi [Citation31]. For example, Wang et al. identified versicoloids A and B from the deep-sea fungus As. versicolor, which shows strong fungicidal effect against Colletotrichum acutatum [Citation32]. Li et al. identified a compound from the culture of a deep-sea fungus As. wentii SD, which exhibits potent inhibitory activities against four plant-pathogenic fungi [Citation33]. Deep-sea fungi have shown much promise in terms of novel and unique secondary metabolites, however, to our knowledge, there has been no report on the antiaflatoxigenic activity of deep-sea fungi.
The main objective of this study was to isolate and characterize the culturable fungi present in deep-sea sediments of the South Atlantic Ocean and examine their antiaflatoxigenic activity.
2. Materials and methods
2.1. Sample collection
Fifteen deep-sea sediment samples used in this study were collected from different sites of the South Atlantic Ocean (). They were collected by SBE-911 plus CTD from a depth of 400–3200 m from July to August 2012 by Cruise DY26 and maintained at 4 °C on board. After transporting the samples to the laboratory, isolation was conducted in October 2012.
Table 1. A list of deep-sea sediment samples with details of their sampling information.
2.2. Media
Four cultivation media and their compositions are as follows: (1) Gause No.1 medium, 20 g of soluble starch, 1 g of KNO3, 0.5 g of NaCl, 0.5 g of K2HPO4, 0.5 g of MgSO4, 0.01 g of FeSO4, 1000 mL of seawater, pH 7.2–7.4; (2) ISP2 medium, 4 g of yeast extract, 10 g of maltose, 4 g of glucose, 1000 mL of seawater, pH 7.2–7.4; (3) seawater PDA medium, 200 g of diced potato was boiled in 1000 mL of boiling seawater for 30 min; 15 g of glucose, 1 g of yeast extract, and 3 g of peptone were added to the filtrate, and the volume was brought to 1000 mL with deionized water; the final pH was adjusted to 6.5; (4) GY medium, 5 g of yeast extract, 20 g of glucose, 1000 mL of deionized water. Twenty grams of agar were added to 1 L of the medium when necessary.
2.3. Isolation of deep-sea fungi
Sediment samples from 15 collection sites were serially diluted up to 10−3 with sterile seawater. One hundred microliters of each dilution was spread-plated on agar media made with seawater. PDA, Gause No.1, and ISP2 media were used for fungal isolation of the TVG15 sediment sample. The Gause No.1 medium was used for fungal isolation from all the sediment samples. The plates were incubated at 28 °C for 1–4 weeks until the morphology of fungi could be distinguished. During the incubation, plates were monitored daily for fungal growth, colony type, and number of colonies. The fungal isolates were picked and transferred to a new corresponding agar plates on the basis of their morphological differences based on visible examination of growth characteristics. Impure strains were streaked on plates until a pure colony with unique culture morphology was observed. Purified fungal cultures were maintained as slant at 4 °C and stored in 20% glycerol at −80 °C, respectively.
2.4. DNA extraction and PCR amplification
DNA from deep-sea fungal cultures was extracted using the method described by Stoeck and Epstein [Citation34]. An aliquot of fresh fungal mycelia (200 mg) was placed in a precooled mortar, fully ground after adding liquid nitrogen three times and transferred to an Eppendorf tube. Six hundred microliters of extraction buffer (100 mM Tris–HCl (pH 8.0), 20 mM Na2EDTA, 0.5 M NaCl, and 1% sodium dodecylsulfate) was added to the tube, incubated in a water bath at 65 °C for 45 min and centrifuged at 12,000 rpm for 5 min. The supernatant was transferred to a new Eppendorf tube, and an equal volume of saturated phenol/CHCl3/isoamyl alcohol was added. The solution was mixed thoroughly and centrifuged for 3 min. The aqueous phase was transferred to a new tube and mixed with 1 vol of isopropanol. Samples were incubated at −20 °C for 30 min and centrifuged at 4 °C for 20 min at 12,000 rpm to recover the precipitate. The pellet was rinsed with 70% EtOH, allowed to air dry briefly and subsequently resuspended in 20 µL of 10xTE (1 mM Tris–HCl, 0.1 mM EDTA, pH 8.0) containing ribonuclease A at 20 µg/mL. One microliter of the DNA was subjected to gel electrophoresis before freezing at −20 °C.
From the extracted fungal DNA, the internal transcribed spacer (ITS) rDNA gene region was amplified by PCR using the primer-pair ITS1 and ITS4 [Citation35]. ITS amplicons were generated for all strains by using the primer pair ITS1-F, 5′-CTTGGTCATTTAGAGGAAGTAA-3′, and ITS4, 5′-TCCTCCGCTTATTGATATGC-3′ [Citation35]. The PCR was performed in 50 µL volumes containing 2 μL of template DNA (∼50 ng/μL), 5 μL of Taq buffer, 1 μL of dNTPs (10 mM), 1 μL of each primer (20 pM) (BGI, Beijing, China) and 4 μL of Taq polymerase (2.5 U/μL). Nuclease-free water was added to increase the volume to 50 µL. The ITS region was amplified at 94 °C for 5 min (initial denaturation), followed by 35 cycles at 94 °C for 1 min (denaturation), 55 °C for 1 min (primer annealing), 72 °C for 2 min (elongation), and a final extension at 72 °C for 10 min. PCR products were purified by a QIAquick PCR purification kit (QIAGEN, Hilden, Germany) following the manufacturer’s protocol. Sequencing was carried out by BGI. Fungal ITS sequences of the 29 isolates were deposited in GenBank under accession numbers MK140679-MK140707.
2.5. Phylogenetic analyses
The ITS sequences obtained were subjected to BLAST search in the NCBI database to determine the sequence similarity. The ITS sequences of closely related fungal strains were retrieved from NCBI. Multiple sequences were automatically aligned using the CLUSTAL W software [Citation36]. Subsequently, phylogenetic analysis of the related ITS sequences was performed using MEGA software version 7.0 [Citation37] with the neighbor-joining [Citation38] method. Bootstrap values were determined based on 1000 replications.
2.6. Liquid fermentation of deep-sea fungi in seawater media
A 1% amount of spore suspension (105) was inoculated into the Gause No.1 media with 100 mL of seawater and incubated at 28 °C and 120 rpm for 7 d. Then, mycelia were separated from the liquid culture by sterile gauze, and the pH of the filtrate was measured. Then the filtrate was centrifuged at 8000×g for 20 min at room temperature, and the supernatant was stored at −20 °C until use. The mycelia were washed with deionized water until the washing liquid became colorless, placed in a 60 °C oven, and dried to constant weight before their fresh weight was measured.
2.7. Determination of antiaflatoxigenic activity by tip culture assay
The tip culture assay was carried out as described previously [Citation39]. Pipette tips were weighed and placed in glass tubes. They were then covered with plastic caps and autoclaved. Before using, the pointed end of the tip was sealed with sterile parafilm in a Clean Bench. The supernatants collected after the liquid fermentation were supplemented with GY (2% glucose and 0.5% yeast extract) to compensate for the consumption of nutrients by deep-sea fungal growth, and the pH of the medium was adjusted to approximately 6.0. After filter sterilization with a 0.22-μm pore-size Millipore membrane, the resulting solution was used for the tip culture assays. An aliquot (700 μL) of the resulting solution and a drop of the spore suspension of NFRI-95 were added into the tip, and incubated at 28 °C for 6 d. Uncultivated media supplemented with GY with and without As. parasiticus NFRI-95 were used as controls for tip culture. Each experiment was carried out in three replicates.
The As. parasiticus mutant strain NFRI-95 was used as an indicator in the antiaflatoxigenic bioassays. It did not produce aflatoxin but accumulated norsolorinic acid, the first stable precursor in the biosynthetic pathway of aflatoxin, in the mycelia. Norsolorinic acid is vivid red and visible to human eyes. The antiaflatoxigenic activity was evaluated in two ways: inhibition of mycelia growth and inhibition of toxin production. For determination of the mycelial growth inhibition ratio, the fresh mycelia weights of the NFRI-95 strain after 6 d’ incubation in the control tip and the experiment tip were measured. To assess the toxin inhibition ratio (suppression of red pigment production), norsolorinic acid in the mycelia of the tip culture after 6 d’ incubation was extracted with a solution containing 1 mol/L NaOH and methanol (1:9, vol/vol). Then, the OD560 nm of the extract was measured. The mycelial growth inhibition ratio and aflatoxin inhibition ratio (or red pigment inhibition ratio) were calculated according to the following formulas:
Mycelial growth inhibition ratio (%)=(W2-W1)/W2 × 100%, where W2 is the fresh weight of the mycelia in the control tip and W1 is the fresh weight of the mycelia in the experiment tip.
Toxin inhibition ratio (%)=(A2-A1)/A2 × 100%, where A2 is the OD560 nm of the control tip and A1 is the OD560 nm of the experiment tip.
3. Results and discussion
3.1. Differences in number and species of deep-sea fungal isolates among different isolation media
There are several ways to cultivate deep-sea fungi, and their efficiency may differ. Damare et al. [Citation18] reported that the percentages of culturable fungi obtained by dilution plating are better than particle plating. We therefore selected the dilution plating method for isolation of the culturable fungi from the sediment samples in this study. Fungi from the deep-sea sediment of the TVG15 sample were isolated from three different media-seawater PDA, Gause No.1, and ISP2 media. The results show that the number and species of fungal isolates were affected by the isolation media. Twelve deep-sea fungal strains were isolated from Gause No.1 medium, while only two strains and one strain were isolated from seawater PDA and ISP2 media, respectively. Taxonomically, cultivation by Gause No.1 medium resulted in the recovery of strains belonging to four species of two genera (Aspergillus and Penicillium), whereas cultivation by the seawater PDA medium resulted in two species of two genera (Aspergillus and Penicillium). Inorganic salts in the Gause No.1 medium probably acted as an important growth factor for the growth of deep-sea fungi. Although only one strain was recovered from ISP2 medium, it was the rarest genus (Cladosporium) isolated from the TVG15 sample. Based on the above results, we selected Gause No.1 medium for the following isolation of culturable fungi in the deep-sea sediments.
3.2. Deep-sea fungal diversity from different sediments
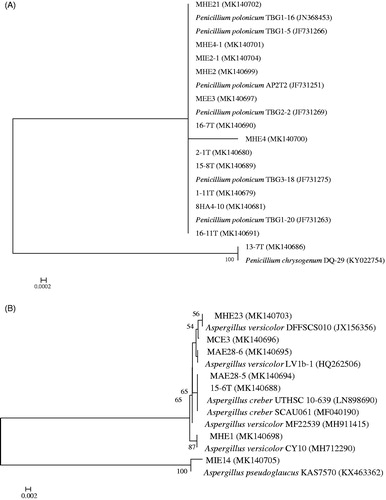
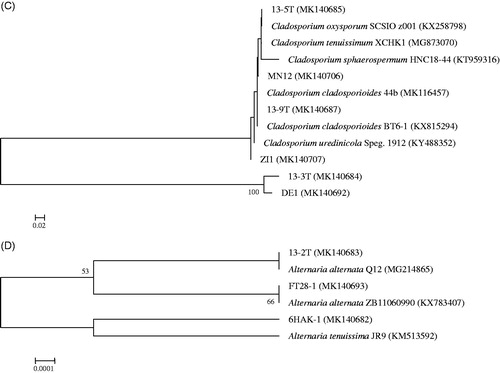
In total, 29 strains were isolated from 15 deep-sea sediments of the South Atlantic Ocean (). The phylogenetic analysis of the ITS sequence of the fungal strains showed that 29 fungal strains all demonstrated ≥98% similarity with sequences from their closest relative species (). The reported Ascomycota isolated from deep-sea sediments mainly belonged to Dothideomycetes, Eurotiomycetes, Leotiomycetes, Saccharomycetes, and Sordariomycetes [Citation12,Citation14,Citation40,Citation41]. In this research, we succeeded in the isolation of deep-sea fungal strains belonging to two classes of the phylum Ascomycota, namely Dothideomycetes, and Eurotiomycetes. As shown in , TVG15 harbored the most fungal strains (15 out of the 29 strains), followed by TVG13 (5 strains) and TVG16 (3 strains). However, only one strain was recovered from TVG01, TVG02, TVG04, TVG06, TVG08, and TVG10. Meanwhile, no strain was recovered from the following six sediments: TVG03, TVG05, TVG09, TVG12, TVG14, and TVG21, although the same methods were used for isolation. The fungal isolates were affiliated with four fungal genera, including Aspergillus, Cladosporium, Penicillium, and Alternaria. Penicillium, accounting for 44.8% of the total fungal strains, was a dominant genus, followed by Aspergillus, Cladosporium, and Alternaria. Fungal taxa distributed unevenly at different sites. Aspergillus could only be isolated from sample TVG15. Some Penicillium, Cladosporium, and Alternaria species were found exclusively in the collection sites TVG04, TVG06, TVG10, or TVG13. Both samples TVG13 and TVG15 harbored five species, which were the most diverse among all 15 sediments. Since the total number of deep-sea fungi isolated was limited, it was hard to show any clear pattern of fungal diversity in relation to the depth of water.
Figure 1. Neighbor-joining phylogenetic trees of fungal strains isolated from the 15 deep-sea sediment samples of the South Atlantic Ocean constructed with ITS rDNA sequences. (A) Penicillium species; (B) Aspergillus species; (C) Cladosporium species; (D) Alternaria species. Bootstrap values (expressed as percentages of 1000 replications) of >50% are shown at branch points.
Table 2. Diversity of the fungi isolated from the 15 deep-sea sediment samples of the South Atlantic Ocean using Gause No. 1 medium.
Among the four genera recovered in our deep-sea sediment samples, Aspergillus sp., Penicillium sp. and Cladosporium sp. have also been isolated in culturable form by many other studies [Citation7,Citation12,Citation23,Citation42]. However, to our knowledge, the presence of Alternaria sp. in the deep-sea was only reported by a research studying the fungal community structure in the deep-sea sediments from the East India Ocean [Citation43]. In their study, Alternaria sp. was not only recovered using traditional cultivation, but also detected by targeted environmental sequencing. Interestingly, we also found their presence in the deep Atlantic Ocean. Most of the fungal isolates in this study exhibited high phylogenetic similarity to terrestrial fungal species, which supports the hypothesis that sedimentation may play an important role in the accumulation of facultative marine fungi in deep-sea sediments [Citation44,Citation45].
In this study, we only succeeded in the isolation of 29 culturable deep-sea filamentous fungi from the 15 deep-sea sediments of the South Atlantic Ocean. This could be attributed to the following reasons. First, about 90% of our isolates belong to Aspergillus sp., Penicillium sp. and Cladosporium sp., and they are reported as fast-growing deep-sea fungi [Citation24], so oligotrophic strains that grow slowly might be ignored because of the culture media chosen in our study. Besides, it has also been reported that cultivation under anoxic conditions may recover fungi that are more divergent from known taxa [Citation40]. So, fungal communities living under anoxic conditions might be missed in our study. Second, it was found that fungi are much more associated with animals rather than mineral substrate [Citation7]. Third, fungi are possibly rare in the deep-sea sediments we sampled. Bass et al. used deep-sea environmental gene libraries, which were constructed based on 11 deep-sea samples from around the world representing depths from 1500 to 4000 m, to study fungal diversity in the deep oceans. Surprisingly, only 18 fungal phylotypes are recovered after sequencing all the clones in the libraries, although the sampling of fungi in their deep-sea libraries is close to saturation [Citation46]. They concluded that fungi are relatively rare in the deep-sea habitats they sampled. In this research, we might have encountered a similar situation as theirs. Last but not least, frozen samples might have an unfavorable effect on culturability.
3.3. Antiaflatoxigenic activity of the deep-sea fungal strains
In order to test the antiaflatoxigenic activity, all 29 fungal isolates were subjected to fermentation in the Gause No.1 liquid media, and their cell-free supernatants were tested against the As. parasiticus mutant strain NFRI-95 by tip culture assay. The results are summarized in . After fermentation, a large proportion of the supernatants had lower pH than their initial pH. The dry weights of the mycelia ranged from 2.41 to 12.69 mg/mL. The antiaflatoxigenic activity was assessed in two ways, inhibition of mycelia growth and inhibition of toxin. Most of the fungal isolates showed moderate to significant antiaflatoxigenic activities, except for two isolates (ZI1 and 15-6 T) which failed to show any antiaflatoxigenic activity. Surprisingly, four strains, MHE21 (P. polonicum), MHE23 (As. versicolor), 13-7 T (P. chrysogenum), and 13-9 T (Cladosporium cladosporioides), could completely inhibit not only mycelial growth, but also aflatoxin production. Three fungal isolates, MN12 (C. cladosporioides), MEE3 (P. polonicum), and MIE14 (A. pseudoglaucus), exhibited 100% toxin inhibition ratio and a mycelial growth inhibition ratio higher than 85%. The three isolates belonging to the genus Alternaria showed moderate antiaflatoxigenic activity. For most of the fungi that displayed strong antiaflatoxigenic activity, the mycelial dry weight was between 4 and 10 mg/mL, but MIE14, which had a mycelial dry weight of merely 2.421 mg/mL, showed a mycelial inhibition ratio of 94% and an aflatoxin inhibition ratio of 100%. On the contrary, MHE1, which had a mycelial dry weight of more than 10 mg/mL, only exhibited weak antiaflatoxigenic activity. This indicated that the antiaflatoxic activity was not related to the mycelial dry weight.
Table 3. Fermentation characteristics of the 29 deep-sea fungi and antiaflatoxigenic activities of the supernatants of their liquid cultures.
Fungi have served as an important source for bioactive secondary metabolites of medicinal and agricultural use. The fungal species which showed potent antiaflatoxigenic activity in our study were also reported to exhibit other antifungal activities. Members of the genus Penicillium are well known for producing a variety of bioactive compounds [Citation47]. It is said that P. polonicum genome encodes 78 biosynthetic gene clusters, the highest among the sequenced Penicillium species [Citation48] and far beyond the number of products that have been discovered till now [Citation49]. The terrestrial fungi P. polonicum isolated from Huperzia serrata was found to exhibit antifungal activity against wilt-inducing fungus Fusarium oxysporum [Citation49]. The marine-derived P. polonicum displays antifungal activity against two plant pathogens Co. acutatum and F. oxysporum, and the marine-derived P. chrysogenum shows antifungal activity against Co. acutatum [Citation50]. Mangrove endophytic fungus P. chrysogenum remarkably inhibits the plant pathogenic fungus Rhizoctonia solani and Co. gloeosporioides [Citation51]. Marine alga-derived P. chrysogenum shows potent inhibitory activity against Aspergillus niger and moderate activity against Alternaria brassicae [Citation52]. However, to our best knowledge, our article is the first time to report the antiaflatoxic activity of Penicillium species. This also suggests that Penicillium species could be promising biological control agents. Similarly, the terrestrial C. cladosporioides was reported to exhibit antifungal activity against plant pathogens Co. acutatum, Co. fragariae, Co. gloeosporioides, and Phomopsis viticola [Citation53]. Jelly-fish associated marine fungi As. versicolor shows antifungal activity against R. solani and Botrytis cinerea [Citation54]. The terrestrial Al. alternata exhibits antifungal activity against Plasmopara viticola [Citation55]. These antifungal activities, together with the antiaflatoxigenic activity reported by us, make the Cladosporium, Aspergillus, and Alternaria species promising biocontrol agents, too. To our knowledge, our study is the first to report the antiaflatoxigenic activity of deep-sea fungi.
In summary, the results presented in this study increase our knowledge and understanding of the diversity of culturable deep-sea fungi in the South Atlantic Ocean. The high proportion of our fungal isolates displayed antiaflatoxigenic activity, which indicates that deep-sea fungi can be of potential use for modern agriculture. The bioactive metabolite(s) that confer the antiaflatoxigenic activity of our fungal isolates need to be identified in the near future.
Disclosure statement
The authors declare no conflict of interest.
Additional information
Funding
References
- Balabanova L, Slepchenko L, Son O, et al. Biotechnology potential of marine fungi degrading plant and algae polymeric substrates. Front Microbiol. 2018;9:1527.
- Xu LJ, Meng W, Cao C, et al. Antibacterial and antifungal compounds from marine fungi. Mar Drugs. 2015;13:3479–3513.
- Moghadamtousi SZ, Nikzad S, Kadir HA, et al. Potential antiviral agents from marine fungi: an overview. Mar Drugs. 2015;13:4520–4538.
- Han WB, Zhang AH, Deng XZ, et al. Curindolizine, an anti-inflammatory agent assembled via Michael addition of pyrrole alkaloids inside fungal cells. Org Lett. 2016;18:1816–1819.
- Morita RY. Starvation survival of heterotrophs in the marine environment. Adv Microbial Ecol. 1982;6:171–198.
- Roth FJ, Orpurt PA, Ahearn DJ. Occurrence and distribution of fungi in a subtropical marine environment. Can J Bot. 1964;42:375–383.
- Burgaud G, Calvez TL, Arzur D, et al. Diversity of culturable marine filamentous fungi from deep-sea hydrothermal vents. Environ Microbiol. 2009;11:1588–1600.
- Nagano Y, Nagahama T, Hatada Y, et al. Fungal diversity in deep-sea sediments-the presence of novel fungal groups. Fungal Ecol. 2010;3:316–325.
- Edgcomb VP, Beaudoin D, Gast R, et al. Marine subsurface eukaryotes: the fungal majority. Environ Microbiol. 2011;13:172–183.
- Lai X, Cao L, Tan H, et al. Fungal communities from methane hydrate-bearing deep-sea marine sediments in South China Sea. ISME J. 2007;1:756–762.
- Nagahama T, Takahashi E, Nagano Y, et al. Molecular evidence that deep-branching fungi are major fungal components in deep-sea methane cold-seep sediments. Environ Microbiol. 2011;13:2359–2370.
- Singh P, Raghukumar C, Meena RM, et al. Fungal diversity in deep-sea sediments revealed by culture-dependent and culture-independent approaches. Fungal Ecol. 2012;5:543–553.
- Zhang XY, Wang GH, Xu XY, et al. Exploring fungal diversity in deep-sea sediments from Okinawa Trough using high-throughput Illumina sequencing. Deep-Sea Res. Part I. 2016;116:99–105.
- Zhang XY, Zhang Y, Xu XY, et al. Diverse deep-sea fungi from the South China Sea and their antimicrobial activity. Curr Microbiol. 2013;67:525–530.
- Picard KT. Coastal marine habitats harbor novel early-diverging fungal diversity. Fungal Ecol. 2017;25:1–13.
- Takami H, Inoue A, Fuji F, et al. Microbial flora in the deepest sea mud of the Mariana Trench. FEMS Microbiol Lett. 1997;152:279–285.
- Raghukumar C, Raghukumar S, Sheelu G, et al. Buried in time: culturable fungi in a deep-sea sediment core from the Chagos Trench, Indian Ocean. Deep-Sea Res Part I. 2004;51:1759–1768.
- Damare S, Raghukumar C, Raghukumar S. Fungi in deep-sea sediments of the central Indian basin. Deep-Sea Res Part I. 2006;53:14–27.
- Singh P, Raghukumar C, Verma P, et al. Fungal community analysis in the deep-sea sediments of the Central Indian Basin by culture-independent approach. Microb Ecol. 2011;61:507–517.
- Thaler AD, Dover CLV, Vilgalys R. Ascomycete phylotypes recovered from a gulf of Mexico methane seep are identical to an uncultured deep-sea fungal clade from the Pacific. Fungal Ecol. 2012;5:270–273.
- Xu W, Luo ZH, Guo SS, et al. Fungal community analysis in the deep-sea sediments of the Pacific Ocean assessed by comparison of ITS, 18S and 28S ribosomal DNA regions. Deep-Sea Res Part I. 2016;109:51–60.
- Xu W, Pang KL, Luo ZH. High fungal diversity and abundance recovered in the deep-sea sediments of the Pacific Ocean. Microb Ecol. 2014;68:688–698.
- Ogaki MB, Coelho LC, Vieira R, et al. Cultivable fungi present in deep-sea sediments of Antarctica: taxonomy, diversity, and bioprospecting of bioactive compounds. Extremophiles. 2020;24:227–238.
- Singh P, Raghukumar C, Verma P, et al. Phylogenetic diversity of culturable fungi from the deep-sea sediments of the Central Indian Basin and their growth characteristics. Fungal Divers. 2010;40:89–102.
- Zain ME. Impact of mycotoxins on humans and animals. J Saudi Chem Soc. 2011;15:129–144.
- Cho KM, Math RK, Hong SY, et al. Iturin produced by Bacillus pumilus HY1 from Korean soybean sauce (Kanjang) inhibits growth of aflatoxin producing fungi. Food Control. 2009;20:402–406.
- Kong Q, Shan S, Liu Q, et al. Biocontrol of Aspergillus flavus on peanut kernels by use of a strain of marine Bacillus megaterium. Int J Food Microbiol. 2010;139:31–35.
- Zhou Y, Wang J, Gao X, et al. Isolation of a novel deep-sea Bacillus circulus strain and uniform design for optimization of its anti-aflatoxigenic bioactive metabolites production. Bioengineered. 2019;10:13–22.
- Park YC, Gunasekera SP, Lopez JV, et al. Metabolites from the marine-derived fungus Chromocleista sp. isolated from a deep-water sediment sample collected in the Gulf of Mexico. J Nat Prod. 2006;69:580–584.
- Daletos G, Ebrahim W, Ancheeva E, et al. Natural products from deep-sea-derived fungi – a new source of novel bioactive compounds? Curr Med Chem. 2018;25:186–207.
- Wang YT, Xue YR, Liu CH. A brief review of bioactive metabolites derived from deep-sea fungi. Mar Drugs. 2015;13:4594–4616.
- Wang J, He W, Huang X, et al. Antifungal new oxepine-containing alkaloids and xanthones from the deep-sea-derived fungus Aspergillus versicolor SCSIO 05879. J Agric Food Chem. 2016;64:2910–2916.
- Li X, Li XD, Li XM, et al. Wentinoids A-F, six new isopimarane diterpenoids from Aspergillus wentii SD-310, a deep-sea sediment derived fungus. RSC Adv. 2017;7:4387–4394.
- Stoeck T, Epstein S. Novel eukaryotic lineages inferred from small-subunit rRNA analyses of oxygen-depleted marine environments. Appl Environ Microbiol. 2003;69:2657–2663.
- White TJ, Bruns TD, Lee SB, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis N, Gelfand D, Sninsky J, et al. editors. PCR-protocols and applications – a laboratory manual. New York (NY): Academic; 1990.
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680.
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874.
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425.
- Yan PS, Song Y, Sakuno E, et al. Cyclo(L-leucyl-L-prolyl) produced by Achromobacter xylosoxidans inhibits aflatoxin production by Aspergillus parasiticus. Appl Environ Microbiol. 2004;70:7466–7473.
- Jebaraj CS, Raghukumar C, Behnke A, et al. Fungal diversity in oxygen-depleted regions of the Arabian sea revealed by targeted environmental sequencing combined with cultivation. FEMS Microbiol Ecol. 2010;71:399–412.
- Calvez TL, Burgaud G, Stéphane M, et al. Fungal diversity in deep-sea hydrothermal ecosystems. Appl Environ Microb. 2009;75:6415–6421.
- Xu W, Guo S, Pang KL, et al. Fungi associated with chimney and sulfide samples from a South Mid-Atlantic Ridge hydrothermal site: distribution, diversity and abundance. Deep Sea Res Part I. 2017;123:48–55.
- Zhang X, Tang G, Xu X, et al. Insights into deep-sea sediment fungal communities from the East Indian Ocean using targeted environmental sequencing combined with traditional cultivation. PLoS One. 2014;9:e109118.
- Shao Z, Sun F. Intracellular sequestration of manganese and phosphorus in a metal-resistant fungus Cladosporium cladosporioides from deep-sea sediment. Extremophiles. 2007;11:435–443.
- Takishita K, Tsuchiya M, Reimer JD, et al. Molecular evidence demonstrating the basidiomycetous fungus Cryptococcus curvatus is the dominant microbial eukaryote in sediment at the Kuroshima Knoll methane seep. Extremophiles. 2006;10:165–169.
- Bass D, Howe A, Brown N, et al. Yeast forms dominate fungal diversity in the deep oceans. Proc Biol Sci. 2007;274:3069–3077.
- Frisvad JC, Smedsgaard J, Larsen TO, et al. Mycotoxins, drugs and other extrolites produced by species in Penicillium subgenus Penicillium. Stud Mycol. 2004;49:201–241.
- Nielsen JC, Grijseels S, Prigent S, et al. Global analysis of biosynthetic gene clusters reveals vast potential of secondary metabolite production in Penicillium species. Nat Microbiol. 2017;2:17044.
- Bai J, Zhang P, Bao G, et al. Imaging mass spectrometry-guided fast identification of antifungal secondary metabolites from Penicillium polonicum. Appl Microbiol Biotechnol. 2018;102:8493–8500.
- Park MS, Fong JJ, Oh SY, et al. Marine-derived Penicillium in Korea: diversity, enzyme activity, and antifungal properties. Antonie Van Leeuwenhoek. 2014;106:331–345.
- Huang S, Chen H, Li W, et al. Bioactive chaetoglobosins from the mangrove endophytic fungus Penicillium chrysogenum. Mar Drugs. 2016;14:172.
- Gao SS, Li XM, Li CS, et al. Penicisteroids a and b, antifungal and cytotoxic polyoxygenated steroids from the marine alga-derived endophytic fungus Penicillium chrysogenum QEN-24S. Bioorg Med Chem Lett. 2011;21:2894–2897.
- Wang X, Radwan MM, Taráwneh AH, et al. Antifungal activity against plant pathogens of metabolites from the endophytic fungus Cladosporium cladosporioides. J Agric Food Chem. 2013;61:4551–4555.
- Yue Y, Yu H, Li R, et al. Exploring the antibacterial and antifungal potential of jellyfish-associated marine fungi by cultivation-dependent approaches. PLoS One. 2015;10:e0144394.
- Musetti R, Vecchione A, Stringher L, et al. Inhibition of sporulation and ultrastructural alterations of grapevine downy mildew by the endophytic fungus Alternaria alternata. Phytopathology. 2006;96:689–698.