Abstract
In May to July 2019, ovate-leaf atractylodes seedling and plant with Damping-off symptoms were observed in farmer field at Sangju and Mungyeong, Korea. Seven fungal isolates have been retrieved from diseased root tissue and identified as Rhizoctonia solani AG-5 based on morphological and molecular characteristics. To the best of our knowledge, this is the first report on damping-off of ovate-leaf atractylodes caused by R. solani AG-5 in South Korea.
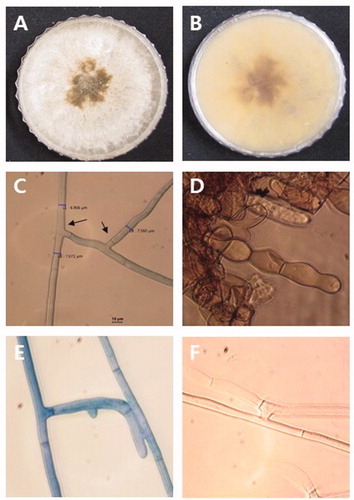
Ovate-leaf atractylodes (Atractylodes ovata De Candolle) is one of the popular medicinal plants in South Korea [Citation1]. Herbs, including ovate-leaf atractylodes (OLA), can be used in different forms, including the use of the whole herb, or use of a ground or powdered form of the raw herb or its dried extracts in teas, syrups, essential oils, ointments, salves, rubs, capsules, and tablets [Citation2,Citation3]. With increases in the demand for OLA, farmers in the Sangju, Mungyeong region in South Korea are using more of their land to cultivate this herb. However, damping-off is a significant threat to the cultivation of OLA. Damping-off leads to substantial economic loss by either affecting direct costs due to damage of the seeds and seedlings or affecting indirect costs due to the replanting of seeds or seedlings [Citation4–6]. In May to July 2019, ovate-leaf atractylodes seedling with damping-off symptoms was observed in farmer field at Sangju and Mungyeong, Korea. Affected seedlings displayed lesions on the stem at the soil line, root rot, and wilt of young seedlings (). The most common fungi and fungal-like organism associated with damping-off include Fusarium spp., Rhizoctonia spp., Pythium spp., Phytophthora spp., and Sclerotium spp. [Citation4–6]. To develop convenient and sustainable damping-off control measures, proper diagnosis of the causal agent is crucial [Citation4]. Seven fungal isolates (three from Sangju and four from Mungyeong) have been retrieved from diseased OLA tissue in an attempt to identify the causative agent associated with damping-off. Small pieces of root excised from the diseased plant, washed with tap water, surface sterilized by dipping in a 0.1% NaOCl solution for 1 min, and then rinsed with double distilled water. Disinfested air-dried root tissue was placed on a petri plate containing water agar (WA). To enhance fungal growth, each WA plate was incubated at 25°C in the dark. After incubation, a 5 mm plug of the emerging hyphal tip from the infected root tissue was transferred to a plate containing potato dextrose agar (PDA, Difco, Becton Dickinson) and incubated at 25°C in the dark. After 20 days of incubation, morphological characteristics of the present isolates, including colony and sclerotia formation, were examined. The colonies were off-white and produced abundant brown sclerotia that were irregular in shape (). Hyphae had a right-angled branching pattern with constriction at the origin of branching (). All the isolates share the same morphological characteristics. The morphological characteristics of the present isolates, including right-angle branching of septate hyphae, brown sclerotia, and brown monilioid cells of sclerotia were consistent with those of Rhizoctonia solani [Citation7].
Figure 1. Damping-off of ovate-leaf atractylodes. (A–B) Symptoms as seen in the field. (C) Symptoms of seedlings inoculated with Rhizoctonia solani AG-5. (D) Healthy seedlings without inoculation.

Figure 2. Microscopic and macroscopic characteristics of Rhizoctonia solani. (A–B) Colony with dark brown sclerotia grown on PDA. (C) Right-angle branching of septate hyphae. (D) Monilioid cells of sclerotia. (E–F) Hyphal anastomosis between a R. solani isolate for anastomosis group designation.
Two isolates were selected for molecular analysis. Approximately 10 grams of mycelium was scraped from a four-day-old actively growing culture of each isolate with a sterilized blade and put into a tube containing 1.5 mL of distilled water. The mycelial mass was homogenized, and the genomic DNA was extracted using the HiGene Genomic DNA Prep Kit (BIOFACT, Daejeon, Korea), as described in the manufacturer’s instructions. Internal transcribed spacers (ITS) region of ribosomal DNA was amplified using the primer set ITS1+ ITS4 [Citation8]. The conditions of PCR amplification were adapted from those described by Hassan et al. [Citation9]. Purification and sequencing of the PCR products were performed commercially by Macrogen, Inc. (Seoul, Korea). The accession numbers (LC569783 and LC569784) of the resulting sequences were obtained after depositing in NCBI GenBank.
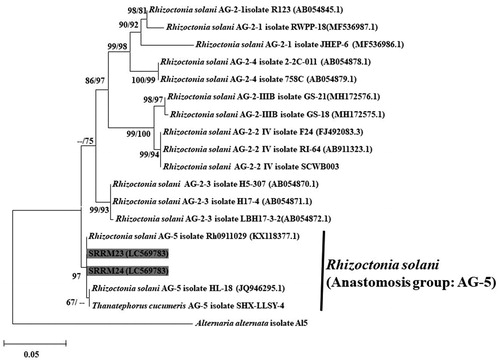
The sequences of the selected molecular marker for each ex-type Rhizoctonia spp. used in the phylogenetic studies performed by Ajayi-Oyetunde and Bradley [Citation7] and Aktaruzzaman et al. [Citation10] were downloaded from GenBank. Sequencing data of ITS region of the isolates used in the present study and the ex-type each fungal species were subjected to multiple sequence alignments with MEGA v. 6.0 using the MUSCLE multiple sequence alignment program [Citation11]. Maximum likelihood (ML) and neighbor-joining (NJ) phylogenetic trees were constructed using MEGA v. 6.0 software [Citation11]. The ITS sequence of the present isolates had 99.45–99.71% similarities to that of R. solani (AG-5 Rh 0911029). ML and NJ trees based on the ITS sequences of the SRRM23 and SRRM24 isolates showed that they formed a clade with R. solani AG-5 and its teleomorph Thanatephorus cucumeris ().
Figure 3. A maximum likelihood tree illustrating estimates of phylogenetic relationship of the Korean and the reference isolates of Rhizoctonia solani. Alternaria alternata isolate Al5 was used as an outgroup. Bootstrap support values ≥50% are presented at the nodes (ML/NJ). The scale bar indicates the number of nucleotide substitutions.
In order to determine the anastomosis group, R. solani AG-2-1 (KACC 40119), AG-2-2-IIIB (KACC 40125), AG-2-2-VI (KACC 40131), AG-3 (KACC 40136), AG-4 (KACC 40140), and AG-5 (KACC 40146) were used as the reference isolates [Citation12]. All the tester strains were obtained from Agricultural Microbiology Division, National Institute of Agricultural Sciences in South Korea. The isolates obtained in the present study and one test strain were grown on a single slide. 5 mm mycelial plugs from the isolates obtained in the present study and one test isolate were placed 1 cm apart on a slide. Each slide containing each pair of fungal plugs was placed in a petri dish containing moist tissue paper and incubated in the dark at 25°C. After seven days of incubation, hyphal anastomosis of was examined using an Olympus BX43 microscope (Tokyo, Japan). The present isolates belong to the AG-5 anastomosis group of R. solani, based on the results of the vegetative compatibility and hyphal fusion test. Hyphae of present isolates fused only with that of AG-5 anastomosis group ().
For the pathogenicity test, OLA seeds were sterilized as described above, and peat soil was sterilized by autoclaving. One hundred grams of autoclaved peat soil was placed in a small pot. Three seeds were sown in each pot, and the pots were incubated at 25°C in a 16-hour light/8-hour dark cycle for germination. 10 g ground mycelium of SRRM23 and SRRM24 mixed with 50 mL of sterile distilled water was poured in the pot. Three pots (containing three same 25 days old size seedlings) were inoculated with single isolate. Three pot inoculated as control with distilled water. After inoculation, all pots were incubated at 25°C with a 16-hour light/8-hour dark cycle in a growth chamber. Wilting of the seedlings was examined regularly. The typical damping-off symptoms were observed in artificially infected seedlings after 10 days of incubation (). The infection rate was estimated as the proportion seedling showing damping-off symptom. The infection rate of R. solani was 90%, while control seedlings remained asymptomatic. In order to confirm Koch’s postulates, the same fungal species were re-isolated from the inoculated seedlings and identified following the procedures described above.
Rhizoctonia solani isolates were recovered from diseased OLA seedlings were identified based on the ITS sequence and morphological characteristics, and in particular the right-angle hyphal branching. Phylogenetic analysis using the ITS sequence of the isolates obtained in the present study and those from the NCBI database verified the results of the morphological analysis. Many studies have used the ITS sequence as a reliable marker for identifying AG and subgroups of Rhizoctonia spp. [Citation7,Citation12]. Rhizoctonia solani (teleomorph: Thanatephorus cucumeris) is a very destructive soil-borne pathogen having wide range of host including soybean, swiss chards and water melon [Citation7,Citation10,Citation12]. To the best of our knowledge there is no report on damping-off of Atractylodes species caused by R. solani. AG-5 is the major group of R. solani has been reported as an important pathogen in a wide range of crops worldwide, including wheat and potato [Citation7,Citation13–15]. In South Korea, R. solani AG-5 has been reported as the causal agent of damping-off of welsh onion (Allium fistulous), ginseng (Panax ginseng), pumpkin (Cucurbita moschata), and cabbage (Brassica oleracea var. capitate) [Citation16]. However, this is the first report of R. solani AG-5 causing damping-off of OLA in South Korea. The result of the present study will help to develop sustainable strategies to control this disease.
Acknowledgements
We would like to thank all of our lab mates for their help during the period of this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Kim JH, Doh EJ, Lee G. Evaluation of medicinal categorization of Atractylodes japonica Koidz. by using internal transcribed spacer sequencing analysis and HPLC fingerprinting combined with statistical tools. Evid Based Complement Alternat Med. 2016;2016:2926819.
- Wachtel-Galor S, Benzie IF. Herbal medicine: biomolecular and clinical aspects. New York, NY, USA: CRC Press; 2011.
- Kim D, Ahn M, Jung J, et al. Perspectives on the market globalization of Korean Herbal Manufacturers: a company-based survey. Evid Based Complement Alternat Med. 2015;2015:1–13.
- Lamichhane JR, Durr C, Schwanck AA, et al. Integrated management of damping-off diseases. Agron Sustain Dev. 2017;37(10):1–25.
- Majeed M, Mir HS, Hassan M, et al. Damping-off in Chilli and its biological management-a review. Int J Curr Microbiol App Sci. 2018;7(04):2175–2185.
- Srivastava JN, Singh AK, Kumar M, et al. Management of damping-off disease of seedling caused in solanaceous and cruciferous vegetable through integrated approach. J Pharmacogn Phytochem. 2018;7(1):3000–3003.
- Ajayi-Oyetunde OO, Bradley CA. Identification and characterization of Rhizoctonia species associated with soybean seedling disease. Plant Dis. 2017;101(4):520–533.
- White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetic. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego (CA): Academic Press; 1990. p. 225–233.
- Hassan O, Lee YS, Chang T. Colletotrichum species associated with Japanese plum (Prunus salicina) Anthracnose in South Korea. Sci Rep. 2019;9(1):12089.
- Aktaruzzaman M, Afroz T, Kim B-S. First report of web blight on Swiss chard caused by Rhizoctonia solani AG2-2 IV in Korea. Plant Dis. 2019;103(12):3287.
- Tamura K, Stecher G, Peterson D, et al. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729.
- Ogoshi A. Ecology and pathogenicity of anastomosis and intraspecific groups of Rhizoctonia Solani Kuhn. Annu Rev Phytopathol. 1987;25(1):125–143.
- Sharon M, Kuninaga S, Hyakumachi M, et al. The advancing identification and classification of Rhizoctonia spp. using molecular and biotechnological methods compared with the classical anastomosis grouping. Mycoscience. 2006;47(6):299–316.
- Woodhall JW, Lees AK, Edwards SG, et al. Characterization of Rhizoctonia solani from potato in Great Britain. Plant Pathol. 2007;56(2):286–295.
- Woodhal JW, Laurenson L, Peters JC. First report of Rhizoctonia solani anastomosis group 5 (AG5) in wheat in the UK. New Dis Rep. 2012;26:9.
- Kim WG, Cho WD, Lee YH. Anastomosis groups and cultural characteristics of Rhizoctonia solani isolates from crops in Korea. Kor J Mycol. 1994;22(4):309–324.
