Abstract
The genus Pseudoperonospora, an obligate biotrophic group of Oomycota, causes the most destructive foliar downy mildew disease on many economically important crops and wild plants. A previously unreported disease by Pseudoperonospora was found on oriental pickling melon (Cucumis melo var. conomon) in Korea, which is a minor crop cultivated in the temperate climate zone of East Asia, including China, Korea, and Japan. Based on molecular phylogenetic and morphological analyses, the causal agent was identified as Pseudoperonospora cubensis, and its pathogenicity has been proven. Importantly, two phylogenetic clades of P. cubensis, harboring probably two distinct species, were detected within the same plots, suggesting simultaneous coexistence of the two clades. This is the first report of P. cubensis causing downy mildew on oriental pickling melon in Korea, and the confirmation of presence of two phylogenetic clades of this pathogen in Korea. Given the high incidence of P. cubensis and high susceptibility of oriental pickling melon to this disease, phytosanitary measures, including rapid diagnosis and effective control management, are urgently required.
Downy mildew (Peronosporaceae; Oomycota) is an obligate biotrophic group that infects a wide range of monocotyledonous and dicotyledonous plants, including many economically relevant crops [Citation1]. The genus Pseudoperonospora is a small group comprising only six species but includes two notoriously pathogenic species, Pseudoperonospora cubensis and Pseudoperonospora humulii. P. cubensis infects many cucurbitaceous crops, such as cucumber, gourd, melon, pumpkin, and watermelon [Citation2,Citation3], and more than 60 host plants have been listed [Citation4,Citation5]. P. humuli is one of the most critical threats to the cultivation of hops (Cannabaceae) [Citation6,Citation7]. Given their association with high economic losses, many recent studies have focused on the biology, host specificity, population structure, detection, and control of Pseudoperonospora species [Citation3,Citation8–13] as well as their taxonomy and phylogeny [Citation14,Citation15].
To date, five Pseudoperonospora species have been reported in Korea, namely P. cannabina, P. cubensis, P. humuli [Citation15–18], P. celtidis [Citation16], and P. urticae [Citation19]. Among them, P. cubensis is the most destructive pathogen on both wild and cultivated Cucurbitaceae in Korea, including Citrullus vulgaris, Cucumis melo, Cucumis sativus, Cucurbita moschata [Citation17], Lagenaria siceraria [Citation20], and Trichosanthes kirilowii [Citation18]. Particularly, P. cubensis raises major economic concerns in the cultivation of C. melo, C. sativus, and C. moschata.
Recently, two phylogenetic clades within P. cubensis, Clades 1 and 2, were described by molecular sequence analysis [Citation14]. Although the clustering showed no clear relevance with the pathotype or geographic distribution [Citation13], Clade 1 seems to be associated with the recent severe epidemics of cucurbit downy mildew in Europe and the United States, while Clade 2 includes pre-epidemic samples originating from East Asia. Interestingly, both clades were often detected in the Czech Republic, Germany, and the USA [Citation13,Citation14]. To date, however, there has been no research to ascertain whether the cucurbit downy mildews in East Asia are caused by two clades, similar to the cases reported in the above countries, or by a particular group, and the identity of the predominant clade remains unclear.
Oriental pickling melon (C. melo var. conomon) is a minor crop cultivated in the temperate climate zone of East Asia, including China, Korea, and Japan. Young fruits are harvested and processed to make salty pickles by putting them in sake lees. From April to June 2020, typical symptoms of downy mildew were observed on the leaves of oriental pickling melon (C. melo var. conomon) growing in Gunsan, Korea (). The disease incidence was high, reaching 30–50% from April to May, but was approximately 90% in June. This study aimed to identify the causal agent of downy mildew on oriental pickling melon in Korea and to determine the causal clade among two phylogenetic clades of P. cubensis, using detailed morphological and multi-locus phylogenetic investigations.
Figure 1. Geographic distribution of Pseudoperonospora cubensis specimens collected on oriental pickling melon (Cucumis melo var. conomon) in Korea. The four plots where P. cubensis specimens were collected are marked on Google Earth map with different colored circles; red for plot 1, blue for plot 2, purple for plot 3, and yellow for plot 4.
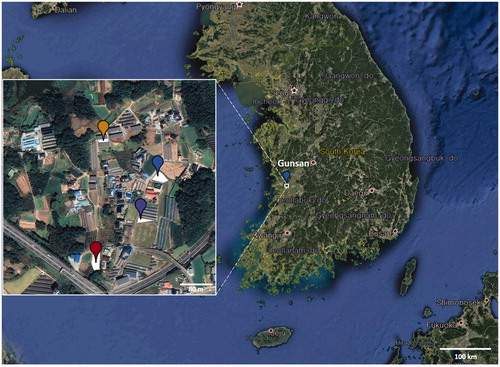
On oriental pickling melon, downy mildew infection resulted in discoloration of the leaf tissues, with yellowish to brownish spots on the upper leaf surfaces ()). The lesions were water-soaked, polyangular, and clearly delimited by the leaf veins (). A distinctive gray to dark brown, dense oomycete growth was observed on the corresponding abaxial leaf surface (). As the disease progressed, the spots turned blackish and often merged to cover larger areas. Several leaves showing downy mildew symptoms were collected within each of four melon plots (116, 164, 780, and 800 m2). displays the site map and a close view of sample locations, which were programmatically marked on Google Earth. All samples were deposited in the Kunsan National University Herbarium (KSNUH), and information on the dried herbarium samples has been provided in .
Figure 2. Downy mildew disease caused by Pseudoperonospora cubensis on oriental pickling melon (Cucumis melo var. conomon) in Korea. (A and B) Downy mildew outbreak in a field of oriental pickling melon; (D and E) Vein-limited spots above (D) and below (E) an infected leaf; (G and H) Close-up view of vein-limited downy mildew growth developing on the lower surface; (J and K) Dense sporangiophores with grayish, numerous sporangia; (C and F) Sporangiophore; (I and L) Sporangium. Scale bars: 100 μm for sporangiophore; 10 μm for sporangium.
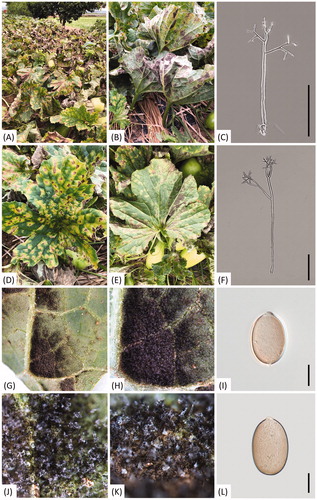
Table 1. Information of Pseudoperonospora collections parasitic to oriental pickling melon (Cucumis melo var. conomon) used in this study.
For a detailed microscopic examination, sporangiophores and sporangia formed underneath the infected leaves were transferred to a drop of lactic acid on a glass slide, covered with a coverslip, and gently warmed using an alcohol lamp. The microscope measurements were examined under the BX53F DIC-light microscope (Olympus, Tokyo, Japan) with the DigiRetina 16M digital camera (Tucsen, Fuzhou, China) or under the Zeiss Imager M2 AX10 microscope (Carl Zeiss, Jena, Germany) with the AxioCam 512 camera (Carl Zeiss, Jena, Germany). Measurements were reported as follows; (minimum–)standard deviation toward the minimum – mean – standard deviation toward the maximum (–maximum). Sporangiophores emerging through the stomata were tree-like, hyaline, straight, or slightly curved, (202.6−)256.3–355.3(−421.9)(av. 305.8) × (4.7−)5.9–8.2(−10.2)(av. 7.1)(n= 100), and monopodially branched in 3–4 orders (). Ultimate branchlets were in pairs, straight to slightly curved (3.6−)6.3–11.6(−15.6)(av. 9.0) µm long, (1.0−)1.3–1.9(−2.5)(av. 1.69) µm wide at the base (n = 100), with a truncate or rarely swollen tip. Sporangia were brownish, ovoidal, or lemon-shaped, measured (19.9−)22.6–27.0(−29.8)(av. 24.8) × (14.3−)15.4–17.9(−19.6)(av. 16.7) µm, with a length/width ratio of (1.30−)1.41–1.57(−1.66)(av. 1.49)(n = 100) (). Resting organs were not observed. Morphological observations revealed that this oomycete unequivocally belonged to the genus Pseudoperonospora, and the characteristics were consistent with those described from P. cubensis (Berk. & Curt.) Rostov. [Citation21].
For molecular sequence analysis, genomic DNA was extracted from the infected plant tissue of the herbarium specimens using the MagListo 5M plant Genomic DNA Extraction Kit (Bioneer, Daejeon, Korea). The mitochondrial cytochrome c oxidase II (cox2) gene was sequenced using primers cox2-F [Citation22] and cox2-RC4 [Citation23]. Amplicons were visualized on 1.2% agarose gel, purified using the AccuPrep PCR Purification Kit (Bioneer, Daejeon, Korea), and sequenced by a DNA sequencing service (Macrogen, Seoul, Korea), with the primers used for amplification. The resulting sequences were edited using the DNASTAR software package version 5.05 (DNASTAR, Madison, WI) and deposited in GenBank (see ). In addition to the reference sequences of Pseudoperonospora species available in NCBI GenBank, they were aligned using MAFFT version 7 [Citation24] with the Q-INS-I algorithm [Citation25]. Minimum evolution (ME) and maximum likelihood trees were constructed with MEGA version 7.0 [Citation26], using the Kimura 2-parameter model and performing 10,000 bootstrap replicates.
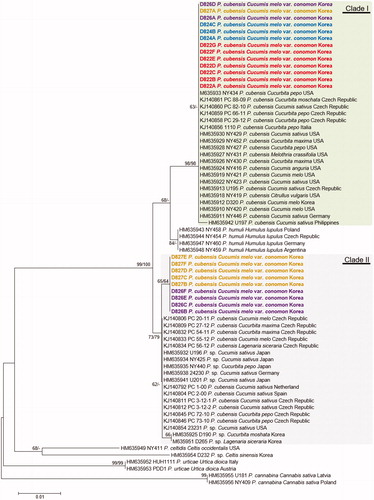
Phylogenetic structures of Pseudoperonospora species, including 22 Korean specimens of oriental pickling, were inferred by the ME and ML methods using cox2 mtDNA sequences, a barcoding locus of oomycetes. As the two trees were congruent, only one ME tree is shown in . In both analyses, the Korean specimens were divided into two different clades, matching each of the two phylogenetic clades of P. cubensis. A sequence dissimilarity of 1.4% (7 out of 472 characters were different) was observed between the two clades. Clade 1 includes Pseudoperonospora specimens of Citrullus, Cucurbita, Cucumis, and Melothria spp. originating from the Czech Republic, Germany, Italy, Korea, the Philippines, and the USA, with a high bootstrapping value of 98/98%. Clade 2 includes Cucurbita, Cucumis, and Lagenaria specimens originating from the Czech Republic, Germany, Japan, Korea, the Netherlands, Spain, and the USA, with a moderate value of 73/79%. Within Clade 2, the Korean specimens formed a subgroup with a weak value of 65/64%. Of the four melon plots of the downy mildew collections, only Clade 1 was detected in plots 1 (n = 7) and 2 (n = 3), Clades 1 and 2 coexisted in the same number in plot 3 (n = 6), and Clade 2 was predominant in plot 4 (n = 6).
Figure 3. Minimum evolution tree of Pseudoperonospora species using cox2 mtDNA sequences. Bootstrapping values (minimum evolution BP/maximum likelihood BP) higher than 60% are shown above the branches (10,000 replicates). The scale bar equals the number of nucleotide substitutions per site. Pseudoperonospora specimens collected in four plots were marked as different colors; red for plot 1, blue for plot 2, purple for plot 3, and yellow for plot 4.
For the pathogenicity test, sporangia were harvested from each infected leaf of two clades (826A in Clade 1 and 826B in Clade 2) collected at site 3. They were suspended in sterile water (1.0 × 106 sporangia/mL) and sprayed onto the lower leaf surface of five healthy C. melo var. conomon. Five control plants were sprayed with distilled water. All plants were incubated in a growth chamber at 22 °C under a relative humidity of 95%. After 7 d, both isolates (826A and 826B) developed downy mildew symptoms on the inoculated leaves. Since there is no difference in disease incidence and characteristics between the two isolates, only the results of the isolate 826A are shown in . It formed yellow to green polygonal lesions on the leaves (), whereas the control plants remained symptomless (). White sporangiophores and dark grayish sporangia were observed on the vein-limited spots of the inoculated leaves (). Their morphological characteristics () and cox2 sequences were consistent with those initially investigated.
Figure 4. Pathogenicity assay of Pseudoperonospora cubensis isolate D826A on oriental pickling melon (Cucumis melo var. conomon). (A) Oriental pickling melon plants a week after inoculation; (B) Uninoculated plants a week after inoculation; (C–E) Vein-limited spots above (C and D) and below (E) an infected leaf; (F and G) Sporangiophores emerging from stomata, with immature sporangia without melanin pigment (F) and mature sporangia with melanin pigment (G).

Based on morphological characteristics, molecular sequencing data, and pathogenicity test, the pathogen was identified as P. cubensis. Previously, P. cubensis has been recorded on C. melo var. conomon in Japan [Citation27] and under laboratory conditions by Zitter et al. [Citation28]. To our knowledge, this is the first report of downy mildew caused by P. cubensis on C. melo var. conomon in Korea. With the recent increase in the popularity of oriental pickling melons, the growing area of this crop in Korea is gradually expanding. However, the high disease severity and low resistance in melons pose a potential risk for oriental pickling melon production.
Beyond the geographic origin and distribution of the two clades of P. cubensis, it should be noted that both clades coexist within a few countries [Citation13,Citation14], including Korea. Additionally, this study has demonstrated that two clades of P. cubensis can coexist even within a small crop plot at the same time. However, it remains unclear whether such a simultaneous coexistence is ubiquitous in other global cucurbit farms, and there is no research on the extent of damage caused by each clade and a disease management method specific to the clade. As the overlapping host ranges and indistinguishable morphological characteristics of the clades pose challenges for their detection and diagnosis in the field and laboratory, there is an urgent need to develop a clade-specific molecular diagnostic method and to study whether they can be distinguished at the species level.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Thines M, Choi YJ. Evolution, diversity, and taxonomy of the Peronosporaceae, with focus on the genus Peronospora. Phytopathology. 2016;106(1):6–18.
- Cohen Y. Downy mildew of cucurbits. In: Spencer DM, editor. The downy mildews. London; New York (NY); San Francisco (CA): Academic Press; 1981. p. 341–354.
- Lebeda A, Cohen Y. Cucurbit downy mildew (Pseudoperonospora cubensis)-biology, ecology, epidemiology, host-pathogen interaction and control. Eur J Plant Pathol. 2011;129(2):157–192.
- Holmes GJ, Main CE, Keever Iii ZT. Cucurbit downy mildew: a unique pathosystem for disease forecasting. In: Spencer-Phillips PTN, Jeger M, editors. Advances in downy mildew research. Vol. 2, Dordrecht: Kluwer Academic; 2004. p. 69–80.
- Lebeda A, Biology and ecology of cucurbit downy mildew. In: Lebeda A, editor. Cucurbit downy mildew. Praha: Czech Scientific Society for Mycology; 1990. p. 13–45.
- Francis SM. Pseudoperonospora humuli. Kew: Commonwealth Mycological Institute; 1983.
- Royle DJ, Kremheller NT. Downy mildew of the hop. In: Spencer DM, editor. The downy mildew. London; New York (NY); San Francisco (CA): Academic Press; 1981. p. 395–419.
- Summers CF, Adair NL, Gent DH, et al. Pseudoperonospora cubensis and P. humuli detection using species-specific probes and high definition melt curve analysis. Can J Plant Pathol. 2015;37(3):315–330.
- Polat I, Baysal O, Mercati F, et al. Characterization of Pseudoperonospora cubensis isolates from Europe and Asia using ISSR and SRAP molecular markers. Eur J Plant Pathol. 2014;139(3):641–653.
- Quesada-Ocampo LM, Granke LL, Olsen J, et al. The genetic structure of Pseudoperonospora cubensis populations. Plant Dis. 2012;96(10):1459–1470.
- Savory EA, Granke LL, Quesada-Ocampo LM, et al. The cucurbit downy mildew pathogen Pseudoperonospora cubensis. Mol Plant Pathol. 2011;12(3):217–226.
- Mitchell MN, Ocamb CM, Grünwald NJ, et al. Genetic and pathogenic relatedness of Pseudoperonospora cubensis and P. humuli. Phytopathology. 2011;101(7):805–818.
- Kitner M, Lebeda A, Sharma R, et al. Coincidence of virulence shifts and population genetic changes of Pseudoperonospora cubensis in the Czech Republic. Plant Pathol. 2015;64(6):1461–1470.
- Runge F, Choi YJ, Thines M. Phylogenetic investigations in the genus Pseudoperonospora reveal overlooked species and cryptic diversity in the P. cubensis species cluster. Eur J Plant Pathol. 2011;129(2):135–146.
- Choi YJ, Hong SB, Shin HD. A re-consideration of Pseudoperonospora cubensis and P. humuli based on molecular and morphological data. Mycol Res. 2005;109(7):841–848.
- Shin HD, Choi YJ. A first check-list of Peronosporaceae from Korea. Mycotaxon. 2003;86:249–267.
- Shin HD, Choi YJ. Peronosporaceae of Korea. Suwon: National Institute of Agricultural Science and Technology; 2006.
- Cho WD, Shin HD. List of plant diseases in Korea. 4th ed. Suwon: Korean Society of Plant Pathology; 2004.
- Choi YJ, Lee HB, Shin HD. Pseudoperonospora urticae occurring on Urtica angustifolia in Korea. Kor J Mycol. 2017;45:160–166.
- Choi YJ, Shin HD. First record of downy mildew caused by Pseudoperonospora cubensis on bottle gourd in Korea. Plant Pathol. 2008;57(2):371–371.
- Waterhouse GM, Brothers MP. The taxonomy of Pseudoperonospora. Mycol Papers. 1981;148:1–18.
- Hudspeth DSS, Nadler SA, Hudspeth MA. COX2 molecular phylogeny of the Peronosporomycetes. Mycologia. 2000;92(4):674–684.
- Choi YJ, Beakes G, Glockling S, et al. Towards a universal barcode of oomycetes-a comparison of the cox1 and cox2 loci. Mol Ecol Resour. 2015;15(6):1275–1288.
- Katoh K, Standley DM. MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780.
- Katoh K, Toh H. Improved accuracy of multiple ncRNA alignment by incorporating structural information into a MAFFT-based framework. BMC Bioinformatics. 2008;9:212.
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874.
- Phytopathological Society of Japan. Common names of plant diseases in Japan. Tokyo: Japan Plant Protection Association; 2019.
- Zitter TA, Hopkins DL, Thomas CE. Compendium of cucurbit diseases. St. Paul (MN): The American Phytopathological Society; 1996.
