Abstract
The Shine Muscat is a table grape, popular in South Korea for its unique mango-flavor taste. Flyspeck is a disease that is characterized by small, black, and circular specks on the grape cuticle was first observed in several commercial orchards in Sangju, South Korea, in August 2019. Here we identified the causal agent of flyspeck based on an advanced diagnosis approach, comprised of both morphological and molecular analyses. Morphological characteristics of the cultures isolated from grape flyspeck were identical to the fungus Cladosporium perangustum. The concatenated sequences of ITS, ACT, and EF1-α were used for molecular phylogenetic analysis, BLAST searches along with Bayesian inference-based phylogeny, confirmed that the causal agent of grape flyspeck is C. perangustum. The cultured fungal isolates also produced flyspeck symptoms on healthy fruits in pathogenicity tests. To the best of my knowledge, this is the first documented evidence of any Cladosporium sp. producing flyspeck symptoms on any plant.
1. Introduction
Shine Muscat is a popular grape variety in South Korea, known for its large, seedless berry, mango flavor, and sweet taste. Because of increasing demand, the market price of Shine Muscat is higher than that of other table grapes. Consequently, more farmers are cultivating Shine Muscat grapes, and production has been increasing since 2016 [Citation1].
However, fungal diseases, such as anthracnose and bunch rot, are a destructive challenge for Shine Muscat cultivation [Citation2,Citation3]. Recently, flyspeck disease was observed at several commercial orchards in Sangju, South Korea. Flyspeck is characterized by small black spots on the grape that resembles fly excreta, which easily rubs off, and is thought to be caused by fungal species that superficially colonize the fruit surface (). Though flyspeck does not cause physical damage to grapes, it greatly reduces marketability, so disease management is critical in orchards. To date, several species of fungi including Zygophiala jamaicensis and Schizothyrium pomi (more than 10 genera) have been associated with flyspeck on different hosts, including apples, pears, carnations, and bananas [Citation4–6]. The identification of the causal agent(s) of flyspeck on grapes is a prerequisite to developing sustainable disease management strategies in fruit orchards. Here we identify the causal agent of this newly emerging disease by employing both morphological and molecular approaches.
Figure 1. Symptoms of flyspeck on Shine Muscat fruits. (A, B) Symptoms on naturally infected grapes; (C, D) Flyspeck symptoms on artificially inoculated fruit.

Shine Muscat fruits showing typical flyspeck symptoms were brought to the laboratory, rinsed with sterile distilled water for one minute, and dried in the hood. Segments of grape peels containing black spots were transferred to water agar (WA) and incubated at 25 °C in the dark. Mycelium growth was visible after one week of incubation. WA plugs containing actively growing mycelia (5 mm in diameter) were then transferred to potato dextrose agar (Difco, Becton Dickinson, Sparks, MD). A total of four isolates were obtained, examined for morphological characteristics, and stored in 10% glycerol at −70 °C.
Fourteen-day-old colonies were examined by microscopy and found to be gray-olivaceous and olivaceous-black when viewed from the reverse side (). Conidiophores were macronematous or semi-macronematous, erect, filiform to cylindrical, 0–5 septate, and 28.4–107.7 μm × 3.0–6.5 μm in size (av. ± SD: 63.1 μm ± 22.5 μm × 4.7 μm ± 0.9 μm). Ramoconidia were cylindrical-oblong, mostly aseptate, with truncate bases and 16.1–33.8 μm × 4.8–5.3 μm (av. ± SD: 22.8 μm ± 5.2 μm × 4.2 μm ± 0.6 μm). The base was 1.6–3.7 μm (av. ± SD: 3.1 μm ± 0.8 μm) in diameter. Secondary ramoconidia were mostly cylindrical, few were oblong and 8.4–20.3 μm × 3.3–4.4 μm (av. ± SD: 13.82 μm ± 2.9 × 3.1 μm ± 0.9 μm). Intercalary conidia were varied in shape, including ellipsoid, ovoid, subcylindrical, and 6.2–15.1 μm × 2.1–4.1 μm (av. ± SD: 9.8 μm ± 1.8 μm × 3.3 μm ± 0.4 μm). Terminal conidia were globose and 2.7–4.4 μm × 1.7–3.8 μm (av. ± SD: 3.7 μm ± 0.5 μm × 2.7 μm ± 0.6 μm) in size (). The morphological characteristics of the flyspeck isolates were almost identical to those of Cladosporium spp., described by Bensch et al. [Citation7] ().
Figure 2. Morphological characteristics of Cladosporium perangustum (SM1). (A, B) Fourteen-day old colony; (C–F) Macronematous conidiophores and conidial chains. Scale bar = 10 μm.
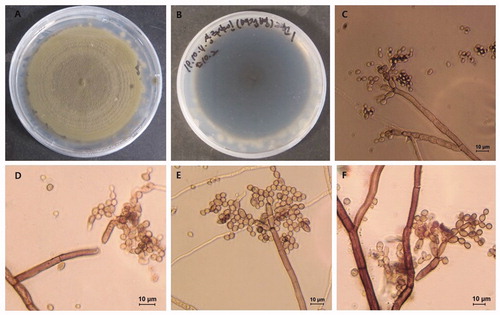
Table 1. Comparative study of morphological characteristics of present and previously identified isolates of Cladosporium perangustum.
For molecular identification, genomic DNA was extracted from seven-day-old colonies of isolates SM1 and SM4 using a HiGeneTM Genomic DNA Prep Kit (BIOFACT, Yuseong-Gu, and Daejeon, Korea). The internal transcribed spacer (ITS), actin (ACT), and translation elongation factor 1- α (EF1-α) genes were targeted for amplification, sequencing, and species identification. Primer pairs ITS1/ITS4, EF1-728F/EF1-986R and ACT-512F/ACT-783R were used to amplify and sequence fragments of ITS, EF1-α, and ACT, respectively [Citation8,Citation9]. The PCR conditions were as follows: an initial denaturation at 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, primer annealing at 53 °C for ITS, 58 °C for ACT and 56 °C for EF1-α for 30 s, primer extension 72 °C for 1 min, and a final extension at 72 °C for 5 min. The success of PCR reactions was confirmed by visualizing in 1% agarose gels (w/v) stained with loading star dye (6×) (Dynebio, Gyeonggi, Korea) in and viewed under a LED illuminator. The confirmed PCR products were purified using the HiGeneTM PCR Purification Kit (Yuseong-Gu, Daejeon, Korea) prior to sequencing. Purified PCR products were sent to Macrogen, Inc. (Seoul, Korea) for sequencing. The generated DNA sequences were analyzed in SeqMan v. 7.1 from the Lasergene package (DNASTAR, Inc. Madison, WI, USA) to obtain consensus sequences and deposited them in GenBank. The GenBank accession number for each gene isolated from the Shine Muscat flyspeck cultures is shown in . The sequences from these isolates were compared to those in the National Center for Biotechnology Information (NCBI) database using BLAST search tools to find the closest fungal species. BLASTn analysis showed that the sequence fragments obtained from the fungal isolates were 99.9, 99.5, and 99.6% homologous to the ITS, EF1- α, and ACT genes from Cladosporium perangustum (strain CBS 125996), respectively. Next, the combined sequences of ITS, EF1-α, and ACT from the fungal isolates, and the reference Cladosporium spp. described by Bensch et al. [Citation7], were used for phylogenetic analysis. Single gene sequences from 33 taxa (Cladosporium spp. reference isolates from Bensch et al. [Citation7]) were aligned using MEGA v. 6.0 [Citation10] and then concatenated with Mesquite v. 2.75 [Citation11]. A 50% majority-rule consensus tree was constructed using MrBayes 3.2.10 [Citation12] and viewed in FigTree v1. 3.1 [Citation13]. The phylogenetic tree shows that genes found in the Shine Muscat flyspeck isolates form a distinct clade with C. perangustum, with high posterior probability ().
Figure 3. A 50% majority-rule consensus tree (Bayesian inference) using a combined dataset of ITS, EF1-a and ACT sequences. The posterior probability (>0.5) are given at the nodes. The scale bar shows the number of substitutions per site.
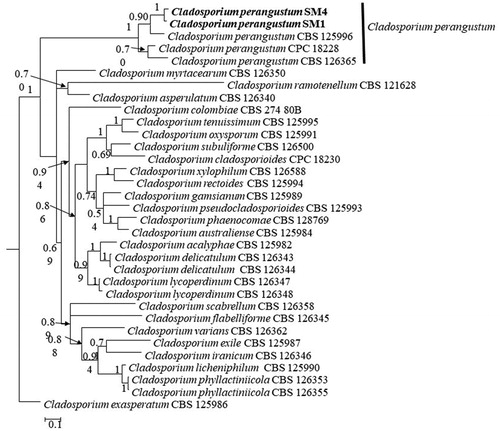
Table 2. List of fungal isolates present and their Genbank accession numbers.
To test C. perangustum pathogenicity on grapes, mature asymptomatic Shine Muscat berries were surface sterilized with 70% ethanol and dried. Spore suspensions were prepared from 25-day old flyspeck cultures. Approximately 15 mL of sterilized distilled water was added to each plate and spores were loosened by scraping the agar surface with a sterile wire loop. The suspension was filtered through Kimtech tissue paper (Kimberly–Clark Professional), and the spore concentration was estimated using a hemocytometer. Healthy fruits were inoculated by swabbing with an inoculum suspension containing 1 × 106 spores mL−1 using a sterilized brush. This process was repeated for each isolate and control berries were inoculated with sterile water alone. Samples were kept in a plastic box and incubated at 25 °C in the dark. This experiment was replicated 12 times, with one berry considered as one replication. At 20 days post-incubation, flyspeck symptoms () were observed on all inoculated berries, identical to the symptoms seen in the orchard, while the control berries remained healthy. Furthermore, flyspeck symptoms were found on 100% of the inoculated berries. The causal agent was re-isolated from the flyspeck and identified as C. perangustum based on morphological characters.
Cladosporium is a common genus of fungi, containing more than 722 named species [Citation14,Citation15]. Bensch et al. [Citation16] revised the taxonomy of Cladosporium and recognized 169 species. Due to the saprophytic and pathogenic nature of fungal species within Cladosporium, this genus has a wide host range [Citation14–16]. Cladosporium sphaerospermum has previously been reported as the causal agent of sooty mold in blueberries, while Cladosporium cladosporioides is the causal agent of the sooty spot on Citrus unshiu (mandarins) [Citation17,Citation18]. This study identified C. perangustum as the causal agent of flyspeck on Shine Muscat grapes. Previously, C. perangustum was implicated in the wood discoloration of pine and larch in South Korea, and leaf spot of Myrica rubra in China [Citation14,Citation19]. The symptoms produced by C. perangustum on grapes are identical to flyspeck. This is the first report of flyspeck being caused by any Cladosporium species anywhere. No Cladosporium species had previously been documented as producing flyspeck symptoms on any plant. Cladosporium spp. are common surface inhabitants on fruit, but have not been associated previously with flyspeck symptoms. Therefore, this report is important not only to alert Korean farmers, crop managers, and plant pathologists about an emerging disease and pathogen but also as an expansion of scientific knowledge of diversity in the SBFS complex, which includes >100 species in many genera.
Acknowledgments
We would like to thank all the member of Plant Pathology Lab, Kyungpook National University for their help.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Korea Rural Economic Institute (KREI). Agriculture in Korea. 2018. Available from: http://www.krei.re.kr/eng/index.do.
- Lim YS, Hassan O, Chang T. First report of anthracnose of shine muscat caused by Colletotrichum fructicola in Korea. Mycobiology. 2020;48(1):75–79.
- Lim Y-S, Hassan O, Kim M-K, et al. First report of bunch rot caused by Aspergillus tubingensis of shine muscat grape in Korea. Plant Dis. 2019;103(11):2953–2953.
- Gao L, Zhang M, Zhao W, et al. Molecular and morphological analysis reveals five new species of Zygophiala associated with flyspeck signs on plant hosts from China. PLoS One. 2014;9(10):e110717.
- Williamson SM, Sutton TB. Sooty blotch and flyspeck of apple: etiology, biology, and control. Plant Dis. 2000;84(7):714–724.
- Batzer JC, Mercedes Diaz Arias M, Harrington TC, et al. Four species of Zygophiala (Schizothyriaceae, Capnodiales) are associated with the sooty blotch and flyspeck complex on apple. Mycologia. 2008;100(2):246–258.
- Bensch K, Groenewald JZ, Dijksterhuis J, et al. Species and ecological diversity within the Cladosporium cladosporioides complex (Davidiellaceae, Capnodiales). Stud Mycol. 2010;67:1–94.
- White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetic. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego (CA): Academic Press; 1990. p. 225–233.
- Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91(3):553–556.
- Tamura K, Stecher G, Peterson D, et al. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30(12):2725–2729.
- Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. Version 2.75; 2011. Available from: http://mesquiteproject.org.
- Ronquist F, Teslenko M, van der Mark P, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542.
- Rambaut A, Drummond A. FigTree v1. 3.1: Tree figure drawing tool. Institute of Evolutionary Biology, Edinburgh, UK, 2009. Available from: http://tree.bio.ed.uk/software/figtree.
- Jang Y, Lee YM, Kim GH, et al. Two species of Cladosporium associated with wood discoloration in Korea. Mycotaxon. 2013;124(1):21–29.
- Dugan FM, Schubert K, Braun U. Check-list of Cladosporium names. Schlechtendalia. 2004;11:1–103.
- Bensch K, Braun U, Groenewald JZ, et al. The genus Cladosporium. Stud Mycol. 2012;72(1):1–401.
- Tashiro N, Noguchi M, Ide Y, et al. Sooty spot caused by Cladosporium cladosporioides in postharvest Satsuma mandarin grown in heated greenhouses. J Gen Plant Pathol. 2013;79(2):158–161.
- Kwon JH, Park K, Lee Y, et al. The occurrence of sooty mold of blueberry caused by Cladosporium sphaerospermum in Korea. J Agirc Life Sci. 2019;53(1):151–156.
- Lu LM, Cheng BP, Pu ZX, et al. First report of Cladosporium perangustum causing leaf spot of Myrica rubra in China. Plant Dis. 2015;99(9):1283–1283.
