Abstract
Polylactic acid (PLA) and polycaprolactone (PCL) are commercially available bioplastics that are exploited worldwide, and both are biodegradable. The PLA and PCL polymer-degrading activity of 30 fungal strains that were isolated from terrestrial environments were screened based on the formation of a clear zone around fungal colonies on agar plates containing emulsified PLA or PCL. Among them, five strains yielded positive results of biodegradation. Strains Korean Agricultural Culture Collection (KACC) 83034BP and KNUF-20-PPH03 exhibited PCL degradation; two other strains, KACC 83035BP and KNUF-20-PDG05, degraded PLA; and the fifth strain, KACC 83036BP, biodegraded both tested plastics. Based on phylogenetic analyses using various combinations of the sequences of internal transcribed spacer (ITS) regions, RPB2, LSU, CAL, and β-TUB genes, the above-mentioned strains were identified as Apiotrichum porosum, Penicillium samsonianum, Talaromyces pinophilus, Purpureocillium lilacinum, and Fusicolla acetilerea, respectively. Based on our knowledge, this is the first report on (i) plastic biodegraders among Apiotrichum and Fusicolla species, (ii) the capability of T. pinophilus to degrade biodegradable plastics, (iii) the biodegradative activity of P. samsonianum against PCL, and (iv) the accurate identification of P. lilacinum as a PLA biodegrader. Further studies should be conducted to determine how the fungal species can be utilized in Korea.
Petrochemical-based plastics are used in large volumes in various industries but cause severe damage to the environment, since they are not degradable [Citation1]. Biodegradable plastics have been developed and commercialized during the past two decades as a potential solution to the problem that arises from the disposal of plastic waste [Citation2]. Synthetic polymers vulnerable to biodegradation include polyesters, polyamides, polyurethanes, poly(amide-enamines), and polyanhydrides [Citation3]. Aliphatic polyesters are biodegradable polymers due to their potentially hydrolyzable ester bonds and are the most representative examples of environmentally friendly polymeric materials [Citation4]. Based on the bonding of constituent monomers, aliphatic polyesters, such as polycaprolactone (PCL), polylactic acid (PLA), and polyglycolic acid (PGA), are classified into polyhydroxyalkanoates [Citation3]. Among them, PLA is the most popular biodegradable polymer, having broad potential applications in agricultural films, biomedical devices, packaging material, and others [Citation5]. Due to its physicochemical properties and compatibility with various polymers, PCL is also considered to be an attractive substitute for nonbiodegradable polymers in commodity applications [Citation6].
Biodegradation of this group of plastics is carried out by fungi, actinomycetes, and bacteria belonging to various taxa. Among known fungi, the members of a few genera were described as degraders of synthetic aliphatic polyesters, including Aspergillus clavatus [Citation7], Fusarium moniliforme [Citation8], Trichoderma viride [Citation9], Tritirachium album [Citation10], and Penicillium oxalicum [Citation11]. The general approach for screening of fungal biodegraders is based upon the selective isolation of polymer-degrading strains from soil, plastic debris, or other environmental samples; however, in several cases, identification of active strains was performed only at the genus level [Citation12–14]. Assessments of already known and deposited fungal species for polymer degrading activities were performed in rare cases, but the studies were usually limited to only few fungal strains [Citation15]. Systematic evaluation of the biodegrading capabilities of a wider range of representatives of various fungal families and genera requires greater research focus to improve our understanding of the biodegradation potential of fungi.
This study was conducted to evaluate 30 fungal strains isolated from terrestrial habitats in Korea based on their ability to degrade synthetic aliphatic polyesters. PLA was selected as a widely used polymer that accounts for 24% of the global production capacity for biodegradable plastics [Citation16]. Considering that degrading capability and rate of biodegradation are directly proportional to the molecular weight of plastics [Citation17], PCL diol was selected as another representative of polyhydroxyalkanoates, as it has a relatively lower molecular weight than that of PLA. PCL diol (m.w. ∼530) was purchased from Sigma–Aldrich (St. Louis, MO, USA), and poly-L-lactic acid was obtained from GFM Co., Ltd. (Suwon, South Korea; ME331050, film, 0.05 mm thickness). Plysurf A210G was purchased from Daiichi Kogyo Seiyaku Co. (Kyoto, Japan). All other compounds were standard commercial preparations that were used without further purification. The search for polymer-biodegradation agents was conducted among fungal strains collected from soil in Korea, all of which were identified at the genus level based on morphological characteristics and phylogenetic analysis (data are not shown). The halo zone technique was used to evaluate the polymer-degrading activity of the selected fungal strains. A mineral medium containing 1% (w/v) PCL or 0.4% emulsified PLA as the sole carbon source was prepared according to previous reports, with only slight modifications [Citation18,Citation19]. The pure colony was transferred onto the plate with minimal salt media supplemented with PLA or PCL, which was solidified with agar. After the incubation period, the polymer-degrading activity of each fungal strain was screened by the formation of a clear (halo) zone around the growing colony on the opaque plate as a result of diffusing enzymes excreted by the fungal hyphae to degrade the plastic. The clear-zone formation test was performed on 30 fungal strains to determine their PCL- and PLA-degrading ability (). In the polymer degradation assays, halo zones were visible around the colonies of the five fungal strains, Korean Agricultural Culture Collection (KACC) 83034BP (=NIBR-WAE-1241), KNUF-20-PPH03, KACC 83035BP (=KNUF-20-PDG06), KNUF-20-PDG05, and KACC 83036BP (=KNUF-20-PBU01) after 7–8 days of incubation. As shown in , strains KACC 83034BP and KNUF-20-PPH03 formed a clear zone around colonies on emulsified PCL plates within 7 days (the halo looks dark due to the black background), which indicates their ability to degrade this polymer. The diameter of the zones increased depending on the incubation time. Two other strains, KACC 83035BP and KNUF-20-PDG05, were incapable of degrading PCL, but both strains formed clear zones around colonies on agar mineral medium supplemented with emulsified PLA. As shows, halo zones were visible after 8 days of incubation; their diameters significantly increased in 18 days, and the polymer was completely degraded after 45 days of incubation. Among the above-mentioned degradative active strains, only KACC 83036BP formed clear zones around colonies on agar medium supplemented with emulsified PLA or PLC, which demonstrated the degradative ability of KACC 83036BP against both polymers (). After at least 4 weeks of incubation, there were no signs of halo zones around the inoculum or growth of the inoculum for the remaining 25 strains in the degradation assays, which indicated that they were incapable of biodegrading PCL and PLA.
Figure 1. Clear zones around the colonies of strain KACC 83034BP (a, b, and c) and KNUF-20-PPH03 (d, e, and f) on a mineral media containing emulsified PCL. (a, d) 7 days after inoculation (DAI); (b, e) 14 DAI; (c, f) 21 DAI. The red arrowhead indicates a clear zone around the colony. The reverse side of the colonies is shown.
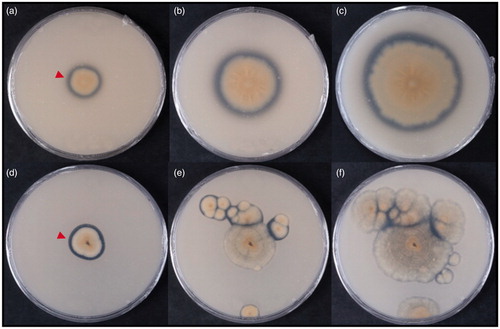
Figure 2. Clear zones around the colonies of strain KACC 83035BP (a, b, and c) and KNUF-20-PDG05 (d, e, and f) on a mineral media containing emulsified PLA. (a, d) 8 days after inoculation (DAI); (b, e) 18 DAI; (c, f) 45 DAI. The red arrowhead indicates a clear zone around the colony. The red arrow indicates PLA particles. The reverse side of the colonies is shown.
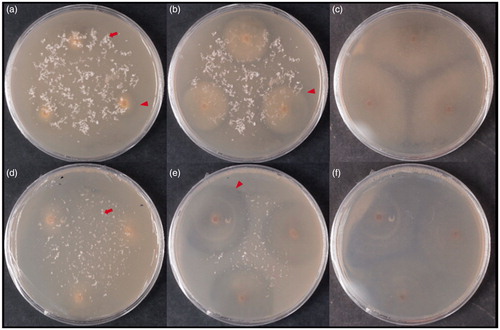
Figure 3. Clear zones around the colonies of strain KACC 83036BP on a mineral media containing emulsified PCL (a, b, and c) and PLA (d, e, and f). (a) 7 days after inoculation (DAI), (b) 14 DAI, (c) 21 DAI, (d) 8 DAI, (e) 18 DAI, (f) 45 DAI. The red arrowhead indicates a clear zone around the colony. The red arrow indicates PLA particles. The reverse side of the colonies is shown.
Table 1. Fungal strains tested for biodegradative activities on PCL and PLA.
The identification of all selected fungal strains at the genus level was previously performed based on their morphological characteristics and phylogenetic analysis using the sequences of the internal transcribed spacer (ITS) regions (data are not shown). A more precise identification at the species level was conducted on the strains that could degrade PCL and PLA. The selected five fungal strains; KACC 83034BP, KNUF-20-PPH03, KACC 83035BP, KNUF-20-PDG05, and KACC 83036BP were cultured on potato dextrose agar (Difco) for 7 days in 25 °C (). Genomic DNA of the five strains was extracted from growing mycelia on PDA using the Solg™ Genomic DNA Prep Kit (Solgent, Daejeon, Korea) following the manufacturer’s instructions. Molecular identification based on polymerase chain reaction (PCR) amplification was conducted using ITS regions, calmodulin (CAL), translation elongation factor 1-alpha (TEF1-α), the large subunit of the nuclear ribosomal RNA (LSU), β-tubulin (β-TUB), and DNA-directed RNA polymerase II second largest subunit (RPB2) genes (separately or in various combinations). The ITS regions of ribosomal DNA (including ITS1, 5.8S, and ITS2) were amplified using the primers ITS1/ITS4 as described by Park et al. [Citation20]. Partial regions of β-TUB, TEF1-α, and CAL genes were amplified using the primer pair T1/Bt2b [Citation21,Citation22], EF1-728F/EF2 [Citation23], and CAL-228F/CAL-737R [Citation23], respectively. The primers LR0R and LR5 [Citation24] were used for LSU amplification, and RPB2 gene was amplified with primers RPB2-5F2 and fRPB2-7cR [Citation25,Citation26]. The amplified PCR products were purified with EXOSAP-IT (Thermo Fisher Scientific, Waltham, MA) and sequenced by Macrogen (Daejeon, Korea). The sequences of allied species were retrieved from the National Center for Biotechnology Information (NCBI) GenBank. Phylogenetic trees were constructed based on concatenated nucleotide sequences of the ITS regions and the partial sequences of selected genes using the neighbor-joining (NJ) method [Citation27], as implemented in MEGA7 [Citation28]. NJ analysis was performed using the Kimura two-parameter distances with gaps excluded from the analysis.
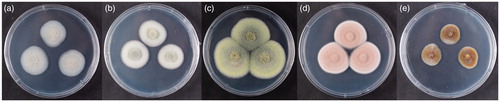
Figure 4. The photographs of selected five fungal strains cultured on PDA for 7 days in 25 °C. (a) KACC 83034BP; (b) KNUF-20-PPH03; (c) KACC 83035BP; (d) KNUF-20-PDG05; (e) KACC 83036BP.
A BLAST search of the NCBI database revealed similarities of 100% between the LSU sequences of KACC 83034BP and Apiotrichum porosum CBS 2040T (NG_068228), A. porosum CBS 8397 (KY106140), and several other strains of this species. A phylogenetic tree based on the LSU gene sequence () confirmed the affiliation of KACC 83034BP with Apiotrichum porosum; this strain was clustered together with the two above-mentioned neighbors in a monophyletic clade with the maximum bootstrap value. The RPB2, CAL, and β-TUB gene sequences of strain KACC 83035BP, respectively shared 100, 99.7–99.8, and 99.7%–99.8% identities with those of Talaromyces pinophilus NRRL 62136 (MH793131, MH793004, MH792940, respectively) and T. pinophilus CBS 631.66T (KM023291, KF741964, JX091381, respectively), and 99.3–99.9% with several other closely related strains of this species. The NJ phylogenetic tree () based on the concatenated β-TUB, CAL, and RPB2 sequences clearly demonstrated that strain KACC 83035BP is identical to Talaromyces pinophilus and that all above-mentioned closest relatives occupy a distinct position from other Talaromyces species. The RPB2 gene region of strain KACC 83036BP revealed a similarity of 100% with that of Fusicolla acetilerea BBA 63789 (HQ897701) but lower than 94% with Fusicolla bharatavarshae PUFD71 (MK157022), Fusicolla betae BBA 64317 (HQ897781), and other Fusicolla species. Based on the ITS region and partial LSU nucleotide sequence similarities, the closest relatives of strain KACC 83036BP were identified as two representatives of Fusicolla acetilerea, strains IMI 181488T (NR_111603) and RF3 (LC333211), which showed 100% similarity. The NJ phylogenetic tree () that was constructed using the concatenated ITS and LSU sequences clearly supported the affiliation of strain KACC 83036BP with Fusicolla acetilerea.
Figure 5. Neighbor-joining phylogenetic tree based on sequences of the LSU gene showing the affiliation of KACC 83034BP with Apiotrichum porosum among the closest Apiotrichum spp. Accession numbers are shown in parentheses. Bootstrap values (based on 1,000 replications) greater than 50% are shown at the branch points. The tree was rooted using Cryptococcus amylolentus CBS 6039T as an outgroup. The isolated strain of this study is indicated in bold. Scale bar = 0.01 substitutions per nucleotide position.
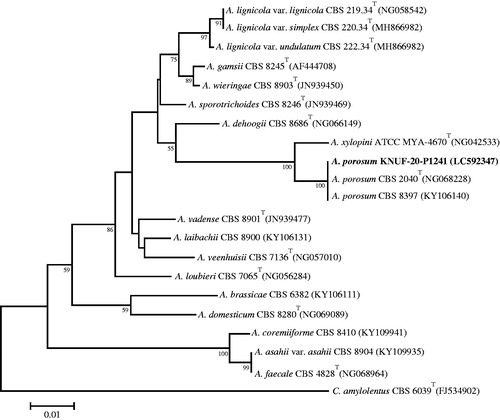
Figure 6. Neighbor-joining phylogenetic tree based on the concatenated sequences of β-TUB, CAL, and RPB2 genes showing the affiliation of KACC 83035BP with Talaromyces pinophilus among the closest Talaromyces spp. Accession numbers of β-TUB, CAL, and RPB2 sequences are, respectively, shown in parentheses. Bootstrap values (based on 1,000 replications) greater than 50% are shown at the branch points. The tree was rooted using Trichocoma paradoxa CBS 788.83 as an outgroup. The isolated strain of this study is indicated in bold. Scale bar = 0.05 substitutions per nucleotide position.
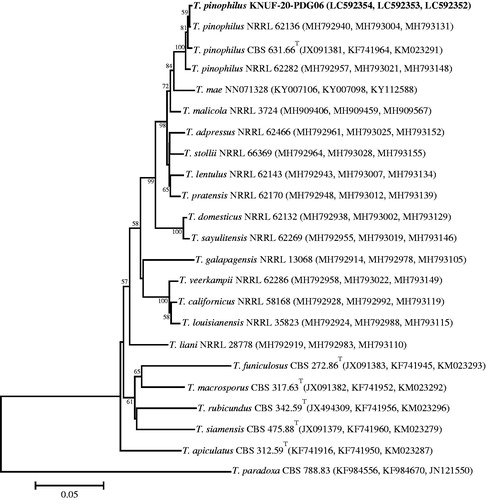
Figure 7. Neighbor-joining phylogenetic tree based on the concatenated sequences of the ITS regions and the LSU gene showing the affiliation of KACC 83036BP with Fusicolla acetilerea among the closest Fusicolla spp. Accession numbers of ITS and LSU sequences are respectively shown in parentheses. Bootstrap values (based on 1000 replications) greater than 80% are shown at branch points. The tree was rooted using Geejayessia celtidicola CBS 125502T as an outgroup. The isolated strain of this study is indicated in bold. Scale bar = 0.01 substitutions per nucleotide position.
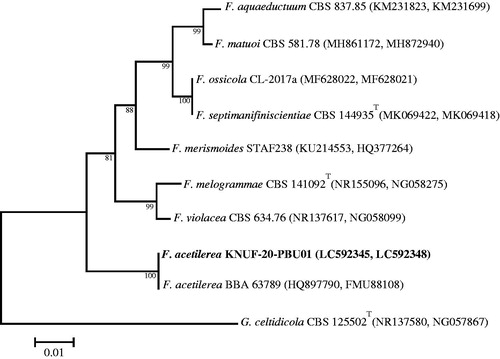
A BLAST search of the NCBI database revealed similarities of 100% between the CAL sequence of KNUF-20-PPH03 and several strains belonging to Penicillium samsonianum, such as AS3.15406 (KJ668583), DTO 327D6 (KT698891), and DTO 187G1 (KT698890). The RPB2 gene region of strain KNUF-20-PPH03 revealed a similarity of 99.73% with that of Penicillium samsonianum AS3.15403 (KT698899) and P. samsonianum DTO 327D6 (KT698901). The β-TUB gene sequence of KNUF-20-PPH03 also demonstrated 99.29%–99.74% identity to several strains of P. samsonianum, including AS3.15406 (KJ668579), PSF373 (MK800137), and SFC100899 (MK682878). The NJ phylogenetic tree () based on the concatenated β-TUB, CAL, and RPB2 sequences showed that strain KNUF-20-PPH03 clustered with representatives of Penicillium samsonianum indicating its affiliation with this fungal species.
Figure 8. Neighbor-joining phylogenetic tree based on the concatenated sequences of β-TUB, CAL, and RPB2 genes showing the affiliation of KNUF-20-PPH03 with Penicillium samsonianum among the closest Penicillium spp. Accession numbers of β-TUB, CAL, and RPB2 sequences, are respectively shown in parentheses. Bootstrap values (based on 1000 replications) greater than 50% are shown at branch points. The tree was rooted using Talaromyces flavus CBS 310.38T as an outgroup. The isolated strain of this study is indicated in bold. Scale bar = 0.05 substitutions per nucleotide position.
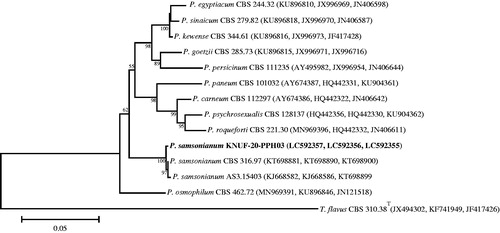
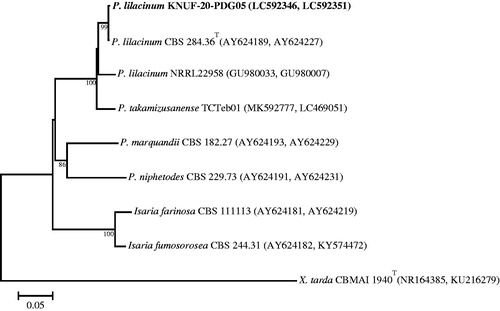
The sequences of the ITS regions and the TEF gene of strain KNUF-20-PDG05 shared 100% identity with those of several strains of Purpureocillium lilacinum, including CBS 128764 (MH865073), IFM 64709 (LC317747), KU20018.18 (MT487832), DTO 39B5 (JF896010), and ER148 (KU738451). In addition, based on the β-TUB gene sequence, KNUF-20-PDG05 had a 99.67% similarity with P. lilacinum CBS 248.36 (JQ965114). The NJ phylogenetic tree () that was constructed using the concatenated ITS and β-TUB sequences clearly supported the affiliation of strain KNUF-20-PDG05 with Purpureocillium lilacinum.
Figure 9. Neighbor-joining phylogenetic tree based on the concatenated sequences of the ITS regions and the β-TUB gene showing the affiliation of KNUF-20-PDG05 with Purpureocillium lilacinum among the closest Purpureocillium spp. and other closely related taxa. Accession numbers of ITS and β-TUB sequences are respectively shown in parentheses. Bootstrap values (based on 1000 replications) greater than 80% are shown at branch points. The tree was rooted using Xenopenidiella tarda CBMAI 1940T as an outgroup. The isolated strain of this study is indicated in bold. Scale bar = 0.05 substitutions per nucleotide position.
In this study, 30 fungal strains were selected and tested for their ability to biodegrade PCL and PLA. Among the fungal species belonging to Ascomycota that are responsible for plastic’s biodegradation, Aspergillus, Fusarium, and Penicillium were mainly encountered [Citation29]. In particular, Penicillium oxalicum DSYD05-1 [Citation11], P. chrysogenum VKM F-227 [Citation15], and P. funiculosum [Citation30] were observed to have the capability to degrade PCL. Based on the above-mentioned results, P. samsonianum KNUF-20-PPH03 is considered to be the novel representative of this genus confirming its ability to degrade PCL. In contrast with the Penicillium species, among members of Apiotrichum, Fusicolla, Talaromyces, and Purpureocillium species, there are only two representatives, namely Talaromyces verruculosus [Citation31] and strain MCM_HumanF8-1, closely related to Purpureocillium lilacinum [Citation32], that were reported to be degraders of polyhydroxybutyrate and poly (butylene succinate), PCL, and PLA, respectively. Our accurate identification of strain KNUF-20-PDG05 as P. lilacinum clearly confirmed the ability of this fungal species to biodegrade PLA. Based on our knowledge, this is the first report of polymer biodegraders among known species of the genera Apiotrichum and Fusicolla and is the first report of Talaromyces pinophilus as a plastic-degrading microorganism. Among the studied fungal species, Fusicolla acetilerea KACC 83036BP, which degraded both PCL and PLA, has a broader spectrum of degradation activity against biodegradable plastics and is considered to be a potential candidate for the treatment of biodegradable plastic wastes. Further research is needed to understand the mechanism of PCL and PLA biodegradation by the five studied strains at the molecular level.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Alimba CG, Faggio C. Microplastics in the marine environment: current trends in environmental pollution and mechanisms of toxicological profile. Environ Toxicol Pharmacol. 2019;68:61–74.
- Pathak VM. Navneet Review on the current status of polymer degradation: a microbial approach. Bioresour Bioprocess. 2017;4:15.
- Vroman I, Tighzert L. Biodegradable polymers. Materials. 2009;2(2):307–344.
- Vert M. Aliphatic polyesters: great degradable polymers that cannot do everything. Biomacromolecules. 2005;6(2):538–546.
- Qi X, Ren Y, Wang X. New advances in the biodegradation of poly(lactic) acid. Int Biodeterior Biodegrad. 2017;117:215–223.
- Fukushima K, Abbate C, Tabuani D, et al. Biodegradation trend of poly(ε-caprolactone) and nanocomposites. Mater Sci Eng. 2010;30(4):566–574.
- Ishii N, Inoue Y, Shimada KI, et al. Fungal degradation of poly (ethylene succinate). Polym Degrad Stab. 2007;92(1):44–52.
- Torres A, Li SM, Roussos S, et al. Screening of microorganisms for biodegradation of poly(lactic-acid) and lactic acid-containing polymers. Appl Environ Microbiol. 1996;62(7):2393–2397.
- Lipsa R, Tudorachi N, Darie-Nita RN, et al. Biodegradation of poly(lactic acid) and some of its based systems with Trichoderma viride. Int J Biol Macromol. 2016;88:515–526.
- Jarerat A, Tokiwa Y. Degradation of poly (L-lactide) by a fungus. Macromol Biosci. 2001;1(4):136–140.
- Li F, Yu D, Lin X, et al. Biodegradation of poly(ε-caprolactone) (PCL) by a new Penicillium oxalicum strain DSYD05-1. World J Microbiol Biotechnol. 2012;28(10):2929–2935.
- Lee K-M, Gimore DF, Huss MJ. Fungal degradation of the bioplastic PHB (poly-3-hydroxybutyric acid). J Polym Environ. 2005;13(3):213–219.
- Masaki K, Kamini NR, Ikeda H, et al. Cutinase-like enzyme from the yeast Cryptococcus sp. strain S-2 hydrolyzes polylactic acid and other biodegradable plastics. Appl Environ Microbiol. 2005;71(11):7548–7550.
- Li F, Hu X, Guo Z, et al. Purification and characterization of a novel poly(butylene succinate)-degrading enzyme from Asperillus sp. XH0501-a. World J Microbiol Biotechnol. 2011;27(11):2591–2596.
- Antipova TV, Zhelifonova VP, Zaitsev KV, et al. Biodegradation of poly-ε-caprolactones and poly-l-lactides by fungi. J Polym Environ. 2018;26(12):4350–4359.
- Haider TP, Volker C, Kramm J, et al. Plastics of the future? The impact of biodegradable polymers on the environment and on society. Angew Chem Int Ed. 2018;57:2–15.
- Muthukumar A, Veerappapillai S. Biodegradation of plastics: a brief review. Int J Pharm Sci Rev Res. 2015;31:204–209.
- Mao H, Liu H, Gao Z, et al. Biodegradation of poly(butylene succinate) by Fusarium sp. FS130 and purification and characterization of poly (butylene succinate). Polym Degrad. 2015;114:1–7.
- Tezuka Y, Ishii N, Kasuya KI, et al. Degradation of poly(ethylene succinate) by mesophilic bacteria. Polym Degrad. 2004;84(1):115–121.
- Park S, Ten L, Lee SY, et al. New recorded species in three genera of the Sordariomycetes in Korea. Mycobiology. 2017;45(2):64–72.
- O'Donnell K, Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol. 1997;7(1):103–116.
- Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 1995;61(4):1323–1330.
- Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91(3):553–556.
- Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172(8):4238–4246.
- Liu YL, Whelen S, Hall BD. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol Biol Evol. 1999;16(12):1799–1808.
- Sung G-H, Sung J-M, Hywel-Jones NL, et al. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, fungi): identification of localized incongruence using a combinational bootstrap approach. Mol Phylogenet Evol. 2007;44(3):1204–1223.
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425.
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874.
- Emadian SM, Onay TT, Demirel B. Biodegradation of bioplastics in natural environments. Waste Manag. 2017;59:526–536.
- Cook WJ, Cameron JA, Bell JP, et al. Scanning electron microscopic visualization of biodegradation of polycaprolactones by fungi. J Polym Sci B Polym Lett Ed. 1981;19(4):159–165.
- Devi SS, Sreenivasulu Y, Rao KVB. Talaromyces verruculosus, a novel marine fungi as a potent polyhydroxybutyrate degrader. Res J Pharm Tech. 2014;7:433–438.
- Penkhrue W, Khanongnuch C, Masaki K, et al. Isolation and screening of biopolymer-degrading microorganisms from northern Thailand. World J Microbiol Biotechnol. 2015;31(9):1431–1442.
