Abstract
The species within the family Cunninghamellaceae are widely distributed and produce important metabolites. Morphological studies along with a molecular phylogeny based on the internal transcribed spacer (ITS) and large subunit (LSU) of ribosomal DNA revealed two new species in this family from soils in China, that is, Absidia ovalispora sp. nov. and Cunninghamella globospora sp. nov. The former is phylogenetically closely related to Absidia koreana, but morphologically differs in sporangiospores, sporangia, sporangiophores, columellae, collars, and rhizoids. The latter is phylogenetically closely related to Cunninghamella intermedia, but morphologically differs in sporangiola and colonies. They were described and illustrated.
1. Introduction
The family Cunninghamellaceae Naumov 1935 ex R.K. Benj. 1959 (Mucorales, Mucoromycetes, Mucoromycota, Fungi) has been under debate on how many genera should be circumscribed. Initially, it included three genera Cunninghamella Matr. 1903, Sigmoideomyces Thaxt. 1891, and Thamnocephalis Blakeslee 1905 [Citation1,Citation2]. Later, the genera Benjaminia S. Ahmad 1967, Chaetocladium Fresen. 1863, Mycotypha Fenner 1932 and Phascolomyces Boedijn 1959 ex Benny & R.K. Benj. 1976 were placed in this family [Citation2–7]. However, Cannon and Kirk [Citation8] and Benny [Citation9] only listed the genus Cunninghamella as its member. Currently, the Encyclopedia of Life curates six genera in this family, that is, Absidia Tiegh. 1878, Chlamydoabsidia Hesseltine & J.J. Ellis 1966, Cunninghamella, Gongronella Ribaldi 1952, Halteromyces Shipton & Schipper 1975, and Hesseltinella H.P. Upadhyay 1970 (http://www.eol.org/, accessed on 16 November 2020). Among these, Absidia and Cunninghamella are commonly encountered.
The species of Absidia are not only potential pathogens for humans and animals [Citation10] but also used to produce 11-α-hydroxylation of medroxyprogesterone and hydrocortisone [Citation11,Citation12]. The genus Absidia possesses stolons, rhizoids, and non-rhizoid-opposite apophysate sporangia with deliquescent walls [Citation13,Citation14], and most species have an apical projection on columellae [Citation15,Citation16]. They are frequently isolated from soil and grow optimally from 20 to 42 °C [Citation17,Citation18]. Currently, Catalogue of Life (http://www.catalogueoflife.org, accessed on 16 November 2020), curates the genus Absidia in Cunninghamellaceae and accommodates 31 species in this genus.
The species of Cunninghamella were usually studied in secondary metabolites, such as polyunsaturated fatty acids [Citation19–21]. They are also mainly isolated from soil and grow optimally from 23 to 28 °C. Traditional morphologies for the classification of this genus include colonies, sporangiophores, vesicles, and sporangiola [Citation22]. It is now comprised of 14 species and three varieties [Citation22,Citation23].
With the development of molecular biology, the rDNA internal transcribed spacer (ITS) has become the DNA barcode for Fungi [Citation24]. However, Absidia was highly variable in ITS sequences [Citation14,Citation17,Citation18]. Consequently, other molecular markers, such as the rDNA large subunit (LSU rDNA) and the gene for actin (act1), were combined with ITS in phylogenetic analyses [Citation17]. For Cunninghamella, the ITS rDNA sequence could effectively identify species [Citation25,Citation26]. Afterward, the LSU rDNA and EF-1α sequences were also applied to reconstruct the evolutionary relationship in Cunninghamella [Citation23,Citation27]. Herein the ITS and LSU rDNA were used for reconstructing the molecular phylogenetic tree.
On the basis of a combination of morphological traits and ITS/LSU rDNA sequences, two new species within the family Cunninghamellaceae in China, each belonging to Absida and Cunninghamella, will be proposed in this study.
2. Materials and methods
2.1. Soil sample collection and strains isolation
Soil samples were collected from Xinjiang (44°43′10′′N, 86°17′03′′E), Beijing (40°33′50′′N, 116°26′26′′E) and Yunnan (102°54′30′′N, 23°42′49′′E) in China. A portion of the soil (1 g) was suspended in sterile water (100 mL) and shaken vigorously. Then, 100 µL of the suspension were spread evenly on a potato dextrose agar (PDA, 20 g/L glucose, 20 g/L agar, 200 g/L potato, and 1000 mL distilled water) plate with antibiotics streptomycin sulfate (100 mg/mL) and ampicillin (100 mg/mL), and incubated darkly at 25 °C. The PDA plate was examined daily with a stereomicroscope (SMZ1500, Nikon, Tokyo, Japan). Upon the presence of colonies, they were transferred to new PDA plates. The living culture was deposited in the China General Microbiological Culture Collection Center, Beijing, China (CGMCC). The dried cultures were deposited in the Herbarium Mycologicum Academiae Sinicae, Beijing, China (HMAS).
2.2. Morphology and growth temperature
Modified synthetic mucor agar (SMA: dextrose 20 g, asparagine 2 g, KH2PO4 0.5 g, MgSO4·H2O 0.25 g, thiamin chloride 0.5 mg, agar 20 g, 1000 mL distilled water, pH7) was used for morphological studies and maximum growth temperature tests [Citation22]. For morphological observation, cultures were incubated at 28 °C for 9 d, and daily examined under a microscope (Axio Imager A2, Carl Zeiss, Oberkochen, Germany); while for determining maximum growth temperature, they were initially incubated at 30 °C for 2 d, and then the incubation temperature increased until the colonies stopped growing. Specifically, a field emission scanning electron microscope (1430VP, Carl Zeiss, Oberkochen, Germany) was used to detect the surface of the sporangia.
2.3. DNA extraction, amplification, and sequencing
Mycelia were grown at 25 °C for 5 d on PDA plates, and then cell DNAs were extracted using a kit (GO-GPLF-400, GeneOnBio Corporation, Changchun, China). The primers NS5M (5′-GGC TTA ATT TGA CTC AAC ACG G-3′) and LR5M (5′-GCT ATC CTG AGG GAA ACT TCG-3′) were used to amplify a fragment covering partial SSU, entire ITS, and partial LSU rDNA [Citation28]. The PCR program was performed with an initial temperature at 95 °C for 5 min, then 30 cycles of denaturation at 95 °C for 60 s, annealing at 55 °C for 45 s and extension at 72 °C for 60 s, and finally an extra extension at 72 °C for 10 min. The PCR product was sequenced with the primer LR5M, as well as ITS4 (5′-TCC TCC GCT TAT TGA TAT GC-3′) and ITS5 (5′-GGA AGT AAA CGT AAC AAG G-3′) [Citation29,Citation30].
2.4. Phylogenetic analyses
ITS and LSU rDNA sequences were used for de novo assembly, manual proofreading, and target extraction with Geneious 8.1 (http://www.geneious.com). For reconstructing a phylogenetic tree, BLAST research was performed in order to retrieve related sequences (). All the sequences were realigned locally using AliView version 3.0 [Citation31]. Phylogenetic analyses were carried out following the methods by Nie et al. [Citation32,Citation33], including maximum-parsimony (MP), maximum-likelihood (ML), and Bayesian Inference (BI) implemented in PAUP version 4.0b10 [Citation34], RAxML version 8 [Citation35] and MrBayes 3.2.7a [Citation36], respectively. MP analyses were conducted using 1000 heuristic search replicates. The best models for the ML and BI analyses were selected with Akaike Information Criterion (AIC) by using jModelTest 2.1.7 [Citation37,Citation38]. ML tree was reconstructed with 1000 bootstrap replicates [Citation35]. In the BI analyses, Markov Chain Monte Carlo (MCMC) chains ran until the convergences met and the standard deviation fell below 0.01. The phylogram was viewed and modified with FigTree version 1.4.4 [Citation39]. Sequence alignments and phylogenetic trees were deposited at TreeBase (submission ID S27294).
Table 1. The taxa of Absidia and Cunninghamella used in phylogenetic analyses.
3. Results
3.1. Taxonomy
Absidia ovalispora H. Zhao & X.Y. Liu sp. nov.
MycoBank No.: 838024
and
Figure 1. Colonies of Absidia ovalispora sp. nov. on SMA at 28 °C in 1–7 d (from left to right). (a) Obverse of CGMCC 3.16018 (ex-holotype strain); (b) Reverse of CGMCC 3.16018 (ex-holotype strain); (c) Obverse of CGMCC3.16019; (d) Reverse of CGMCC3.16019.
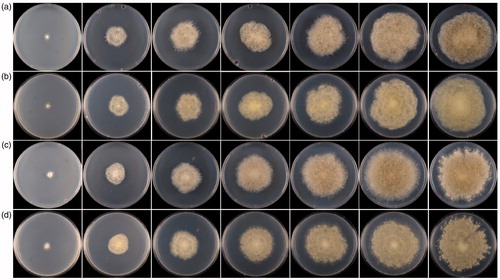
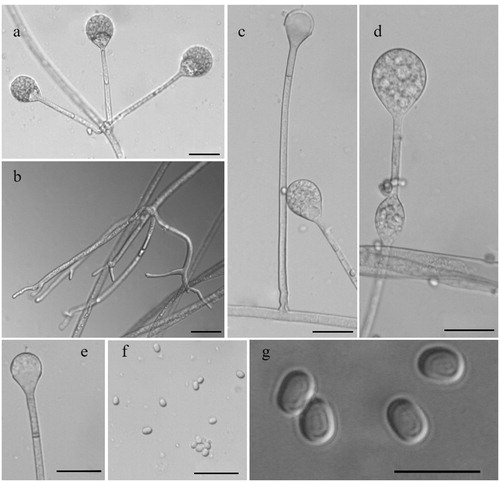
Figure 2. Morphology of Absidia ovalispora sp. nov. CGMCC 3.16018 (ex-holotype strain). (a) Verticillately branched sporangiophores; (b) Rhizoids; (c) Single sporangiospore; (d) Swelling on sporangiosphores; (e) Columella with a septum and an apical projection; (f, g) Sporangiospores. Scale bars: (a, c–f) = 20 μm; (b) = 50 μm; (g) = 10 μm.
Typification: CHINA, YUNNAN: Jianshui County, Honghe Hani and Yi Autonomous Prefecture, 102°54′30′′N, 23°42′49′′E, from soil sample, December 28, 2018, Min Qiao (holotype HMAS 249157, living ex-holotype culture CGMCC 3.16018. GenBank: ITS = MW264130; LSU = MW264071)
Additional culture examined: CHINA, YUNNAN: Jianshui County, Honghe Hani and Yi Autonomous Prefecture, 102°54′30′′N, 23°42′49′′E, from a soil sample, December 28, 2018, Min Qiao (HMAS 249158, living culture CGMCC 3.16019. GenBank: ITS = MW264131; LSU = MW264072).
Etymology: ovalispora (Lat.), referring to the shape of the sporangiospores.
Ecology and distribution: Soil in Yunnan Province, China.
Morphological characteristics: Colonies on SMA reaching 9 cm at 28 °C for 7 d, floccose, initially white, quickly grayish-white, and finally brown, irregular at the edge. Hyphae flourishing. Rhizoids always simple and fingerlike. Stolons present. Sporangiophores on stolons, erect, single or 2–6 verticillate, 27.96–212.96 µm long and 2.14–6.39 µm wide, occasionally septate or swollen 20–30 µm below the sporangia. Apophyses 2.14–14.92 µm long and 5.15–23.94 µm wide. Collars absent. Columellae globose to ellipsoidal, 7.99–20.80 µm long and 7.01–23.88 µm wide, always with an apical projection. Sporangia globose to ellipsoidal, 12.54–40.80 µm long and 12.47–38.54 µm wide. Sporangiospores ovoidal to ellipsoidal, 2.13–5.38 µm long and 1.73–4.57 µm wide. Chlamydospores not observed. Zygospores not observed.
Maximum growth temperature: 37 °C.
Note: On the medium SMA, Absidia ovalispora is distinguished from its allied species Absidia koreana H.B. Lee, H.W. Lee & T.T.T. Nguyen 2015 in sporangiospores, sporangia, sporangiophores, columellae, collars, and rhizoids. Sporangiospores are ovoidal to ellipsoidal in A. ovalispora, while short-cylindrical or cylindrical in A. koreana [Citation40]. Sporangia/sporangiophores/columellae in A. ovalispora (12.54–40.80 µm × 12.47–38.54 µm/2.14–6.39 µm wide/7.99–20.80 µm × 7.01–23.88 µm) are all bigger than those in A. koreana (19.33–23.64 µm × 21.06–26.35 µm/3.84–4.60 µm wide/10.90–16.96 µm × 11.46–18.89 µm) [Citation40]. Collars are absent in A. ovalispora but present in A. koreana [Citation40]. Rhizoids are always observed in A. ovalispora but were not described in A. koreana [Citation40]. Apart from these differences, they are similar in the branching manner, that is, single or up to 6 sporangiophores are arising from the same point on the stolons. This paper reported two strains in the A. ovalispora. It is worth noting the difference in colonies between these two strains, that is, the strain CGMCC 3.16019 is lighter, more vigorous than the ex-holotype strain CGMCC 3.16018. Moreover, CGMCC 3.16019 extends some small satellite colonies around the main colony ().
Cunninghamella globospora H. Zhao & X.Y. Liu sp. nov.
MycoBank No.: 838023
and
Figure 3. Colonies of Cunninghamella intermedia CBS 347.69 and C. globospora sp. nov. CGMCC 3.16020 (ex-holotype strain) on SMA at 28 °C in 1–5 d. (a) Obverse of CBS 347.69; (b) Reverse of CBS 347.69; (c) Obverse of CGMCC 3.16020; (d) Reverse of CGMCC 3.16020.
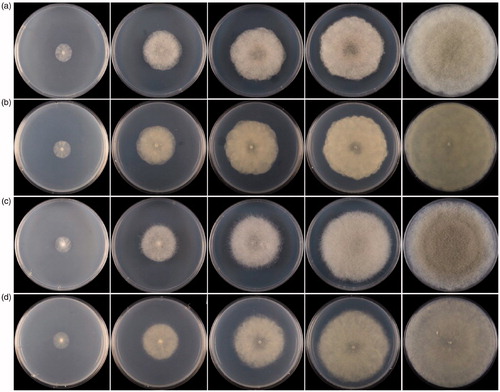
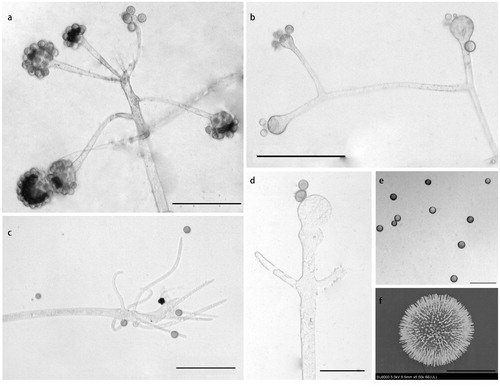
Figure 4. Morphology of Cunninghamella globospora sp. nov. CGMCC 3.16020 (ex-holotype strain). (a, b) Sporophores showing characteristic branching patterns; (c) Rhizoids; (d–f) Sporangiola. Scale bars: (a–c) = 100 μm; (d, e) = 50 μm; (f) = 10 μm.
Typification: CHINA, XINJIANG: Manas County, 44°43′10′′N, 86°17′03′′E, from the soil sample in salt marshes, July 1, 2015, Jing Zhu, (holotype HMAS 249159, living ex-holotype culture CGMCC 3.16020. GenBank: ITS = MW264132; LSU = MW264073.)
Additional culture examined: CHINA, BEIJING: Yanqing County, 40°33′50′′N, 116°26′26′′E, from a soil sample, November 13 2020, Tong-Kai Zong, (HMAS 249160, living culture CGMCC 3.16021. GenBank: ITS = MW264133; LSU = MW264074).
Etymology: globospora (Lat.), referring to the shape of the sporangiospores.
Ecology and distribution: Soil in Xinjiang and Beijing, China.
Morphological characteristics: Colonies on SMA floccose, reaching 9 cm in 5 d at 28 °C, initially white, soon gray, finally gray-brownish, and gray reversely. Hyphae branched, 4.34–18.00 µm diameter, and septate when old. Stolons present. Rhizoids simple, and fingerlike. Sporophores erect or recumbent, arising from stolons and aerial hyphae, main axes usually aequilate and 4.79–24.85 µm diameter, rarely gradually thicken upwards, primary branches 37.85–459.72 µm long and 2.86–13.74 µm wide, re-branching or third-branching into branchlets. Vesicles hyaline to brownish, globose or subglobose, sometimes expanded at the bottom, bearing lots of pedicels which are left after sporangiola fall off, axial vesicles 19.04–43.62 µm diameter, and lateral vesicles 13.74–27.46 µm diameter. Pedicels 1.00–3.07 µm long. Sporangiola globose, 9.09–14.73 µm diameter, and growing on vesicles with pedicels. Chlamydospores absent. Zygospores absent.
Maximum growth temperature: 46 °C.
Notes: On the medium SMA at 28 °C, the Cunninghamella globospora is distinguished from its closely related species Cunninghamella intermedia K.B. Deshp. & Mantri 1966 by sporangiola and colonies. The species C. intermedia was nominated with the sporangiola being intermediate among four species [Citation41]. The sizes of sporangiola in the new species C. globospora (9.09–14.73 µm diameter) are smaller and more uniform than those in C. intermedia (8.00–23.00 µm diameter) [Citation22]. And, colonies are gray-brownish in C. globospora but “Hair Brown” in C. intermedia [Citation22]. Except for these discrepancies, they are similar in other characteristics, such as branched and septate hyphae, fingerlike rhizoids, and long primary branches [Citation22].
3.2. Phylogenetic analyses
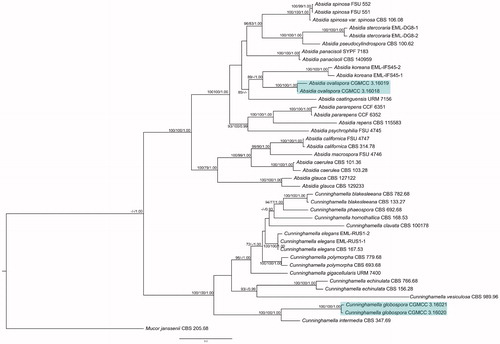
The combined ITS and LSU rDNA dataset consisted of 42 taxa representing 26 species, and 2108 characters including 811 constant, 303 parsimony-uninformative, and 994 parsimony-informative. The generated MP tree length (TL), consistency index (CI), homoplasy index (HI), retention index (RI) and rescaled consistency index (RC) were 5227, 0.4534, 0.5466, 0.6629 and 0.3006, respectively. The optimal model for the BI inference was GTR + I + G, and the final optimization likelihood value is −22601. When convergent, and the average standard deviation of split frequencies was 0.008871. The topologies of the three phylogenetic trees were similar. Hence, the ML tree was selected to represent the evolutionary history, being supported with ML bootstrap, MP bootstrap, and BI posterior probability values (). Absidia ovalispora was closely related to A. koreana (ML = 89, MP = 36, BI = 1.00); and Cunninghamella globospora was closely related to C. intermedia (ML = 100, MP = 100, BI = 1.00).
Figure 5. Maximum-likelihood phylogenetic tree of Absidia and Cunninghamella based on ITS and LSU rDNA sequences, with Mucor janssenii as outgroup. Two new species A. ovalispora sp. nov. and C. globospora sp. nov. are in shade. Maximum-parsimony (MP) bootstrap values (≥70%)/maximum-likelihood (ML) bootstrap values (≥70%)/Bayesian Inference (BI) posterior probabilities (≥0.95) of each clade are indicated along branches. Scale bar indicates substitutions per site. “T” indicates the ex-holotype cultures.
4. Discussion
The genus Absidia is characterized by sporangiophores originating from stolons, apophysate sporangia with deliquescent walls, septa usually seen below sporangia, and heterothallic zygospores with appendages [Citation9,Citation17,Citation40]. The species of Absidia were always isolated from soils and animal dung [Citation42]. In this study the new species of Absidia ovalispra also possesses these morphological features and was isolated from soil in Xinjiang, China.
In the genus Cunninghamella, stable traits used for identification are sporangia, zygospores, maximum growth temperatures, mating compatibilities, and ITS rDNA sequences [Citation22]. According to this criterion, Cunninghamella bigelovii, Cunninghamella gigacellularis, and Cunninghamella guizhouensis were recently proposed [Citation23,Citation27,Citation43]. In this paper, C. globospora was proposed from China based on molecular data and morphological observation.
The phylogenetic analyses on ITS and LSU rDNA () showed that A. ovalispora and C. globospora were closely related to A. koreana (ML = 89, MP = 36, BI = 1.00) and C. intermedia (ML = 100, MP = 100, BI = 1.00), respectively. Their differences in morphology were compared in the respective Notes part. Apart from these, the two new species were both distinct from their allies in the shape of sporangiospores, and thus they were nominated. The maximum growth temperature of C. intermedia was 43 °C in a previous study [Citation22], but 46 °C in this study, same as in the new species C. glaobospora. It is maybe because that the strains are different, IMI 200623 in Zheng and Chen [Citation22] and CBS 347.69 in this study.
With the proposal of these two species of Cunninghamellaceae, the species and variety number of Absidia and Cunninghamella are now 32 and 18, respectively.
Author contributions
Heng zhao, Jing Zhu, Zhi-Dong Zhang, and Xiao-Yong Liu conceived and designed this research. Heng Zhao analyzed data. Xiao-Yong Liu improved the manuscript. Tong-Kai Zong, Qing Lin, and Min Qiao collected and isolated samples. Other authors conducted part experiments and advised on analyses.
Acknowledgments
Ze-Fen Yu (Yunnan University) is acknowledged for helping with specimen collection.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The datasets generated during the current study are available in the TreeBASE repository at https://treebase.org/.
Additional information
Funding
References
- Naumov NA. Opredelitel Mukorovykh (Mucorales). Ed. 2. Moscow (Russia); Leningrad (Russia): Bot. Inst. Acad. Sci. U.S.S.R; 1935. p. 136.
- Benjamin RK. The merosporangiferous Mucorales. Aliso. 1959;4(2):321–433.
- Hesseltine CW. Genera of Mucorales with notes on their synonymy. Mycologia. 1955;47(3):344–363.
- Pidoplichko NM, Mil’ko AA. Atlas mukoral’vykh gribov Atlas of the Mucorales. Izdat. Kiev (Ukraine): ‘Naukova Dumka’; 1971. p. 115.
- Hesseltine CW, Ellis JJ. Mucorales. In Ainsworth GC, Sparrow FK, Sussman AS, editors. The fungi. Vol. 4b. New York (NY): Academic Press; 1955. p. 187–217.
- Mil’ko AA. Opredeltiel’ mukoral’nykh gribov Key to the identification of Mucorales. Kiev (Ukraine): ‘Naukova Dumka’; 1974. p. 303.
- Benny GL, Benjamin RK. Observations on Thamnidiaceae (Mucorales). II. Chaetocladium, Cokeromyces, Mycotypha, and Phascolomyces. Aliso. 1976;8(4):391–424.
- Cannon PF, Kirk PM. Fungal families of the world. Wallingford (UK): CAB International; 2007. p. 456.
- Benny GL. Zygomycetes [Internet]. 2020 [cited Accessed 2020 November 20]. Retrieved from: www.zygomycetes.org
- Eucker J, Sezer O, Graf B, et al. Mucormycoses. Mycoses. 2001;44(7–8):253–260.
- Ghasemi S, Heidary M, Habibi Z. The 11α-hydroxylation of medroxyprogesterone acetate by Absidia griseolla var. igachii and Acremonium chrysogenum. Steroids. 2019;149:108427.
- Chen J, Fan F, Qu G, et al. Identification of Absidia orchidis steroid 11β-hydroxylation system and its application in engineering Saccharomyces cerevisiae for one-step biotransformation to produce hydrocortisone. Metab Eng. 2020;57:31–42.
- van Tieghem P. Troisieme memoire sur les Mucorinees. Annales Des Siences Naturelles, Botanique, Ser VI. 1876;4:312–398.
- Zhang TY, Yu Y, Zhu H, et al. Absidia panacisoli sp. nov., isolated from rhizosphere of Panax notoginseng. Int J Syst Evol Microbiol. 2018;68(8):2468–2472.
- Ling Y. Étude biologique des phenomenes de la sexualite chez les Mucorinees. Appendice Revue Generale de Botanique. 1930;42:722–752.
- Hesseltine CW, Ellis JJ. Notes on Mucorales, especially Absidia. Mycologia. 1961;53(4):406–426.
- Hoffmann K, Discher S, Voigt K. Revision of the genus Absidia (Mucorales, Zygomycetes) based on physiological, phylogenetic, and morphological characters; thermotolerant Absidia spp. form a coherent group, Mycocladiaceae fam. nov. Mycol Res. 2007;111(10):1169–1183.
- Walther G, Pawłowska J, Alastruey-Izquierdo A, et al. DNA barcoding in Mucorales: an inventory of biodiversity. Persoonia. 2013;30:11–47.
- Fakas S, Papanikolaou S, Galiotou-Panayotou M, et al. Organic nitrogen of tomato waste hydrolysate enhances glucose uptake and lipid accumulation in Cunninghamella echinulata. J Appl Microbiol. 2008;105(4):1062–1070.
- Alakhras R, Bellou S, Fotaki G, et al. Fatty acid lithium salts from Cunninghamella echinulata have cytotoxic and genotoxic effects on HL-60 human leukemia cells. Eng Life Sci. 2015;15(2):243–253.
- Zhao H, Lv ML, Liu Z, et al. High-yield oleaginous fungi and high-value microbial lipid resources from Mucoromycota. BioEnergy Res. 2020;
- Zheng RY, Chen GQ. A monograph of Cunninghamella. Mycotaxon. 2001;80:1–75.
- Zhang ZY, Zhao YX, Shen X, et al. Molecular phylogeny and morphology of Cunninghamella guizhouensis sp. nov. (Cunninghamellaceae, Mucorales), from soil in Guizhou, China. Phytotaxa. 2020;455(1):31–39.
- Schoch CL, Seifert KA, Huhndorf S, et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci USA. 2012;109(16):6241–6246.
- Su YC, Huang H, Liu XY, et al. Systematic relationship of several controversial Cunninghamella taxa inferred from sequence comparisons of ITS2 of rDNA. Mycol Res. 1999;103(7):805–810.
- Liu XY, Huang H, Zheng RY. Relationships within Cunninghamella based on sequence analysis of ITS rDNA. Mycotaxon. 2001;80:77–95.
- Guo J, Wang H, Liu D, et al. Isolation of Cunninghamella bigelovii sp. nov. CGMCC 8094 as a new endophytic oleaginous fungus from Salicornia bigelovii. Mycol Prog. 2015;14(3):11.
- Du P, Wu F, Tian XM. Three new species of Junghuhnia (Polyporales, Basidiomycota) from China. MycoKeys. 2020;72:1–16.
- White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gefand DH, Sninsky JJ, et al., editors. PCR protocols: a guide to methods and applications. San Diego (CA): Academic Press; 1990. p. 315–322.
- Wang CG, Liu SL, Wu F. Two new species of Perenniporia (Polyporales, Basidiomycota). MycoKeys. 2020;69:53–69.
- Larsson A. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics. 2014;30(22):3276–3278.
- Nie Y, Cai Y, Gao Y, et al. Three new species of Conidiobolus sensu stricto from plant debris in eastern China. MycoKeys. 2020;73:133–149.
- Nie Y, Yu DS, Wang CF, et al. A taxonomic revision of the genus Conidiobolus (Ancylistaceae, Entomophthorales): four clades including three new genera. MycoKeys. 2020;66:55–81.
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (*and other methods). Version 4.0b10. Sunderland (UK): Sinauer Associates; 2002.
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313.
- Ronquist F, Teslenko M, van der Mark P, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542.
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704.
- Darriba D, Taboada GL, Doallo R, et al. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9(8):772.
- Rambaut A. FigTree version 1.4.4 [Internet]. 2012. Retrieved from: http://tree.bio.ed.ac.uk/software/figtree/
- Ariyawansa HA, Hyde KD, Jayasiri SC, et al. Fungal diversity notes 111–252: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2015;75(1):27–274.
- Deshpande KB, Mantri JM. A new species of Cunninghamella from India. Mycopathologia et Mycologia Applicata. 1966;28(4):342–344.
- Richardson M. The ecology of the Zygomycetes and its impact on environmental exposure. Clin Microbiol Infec. 2009;15:2–9.
- Hyde KD, Hongsanan S, Jeewon R, et al. Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016;80(1):1–270.
