Abstract
Two Diaporthe species isolated from fruit of Citrus sinensis in China were characterized based on morphology and multilocus phylogeny of ITS, tef1, and tub2 gene sequences. The phylogeny indicated that the two species match Diaporthe taoicola and D. siamensis. A critical examination of phenotypic characteristics confirmed the phylogenetic results. Diaporthe taoicola was morphologically characterized by producing Alpha conidia with tapering toward both ends. Meanwhile, D. siamensis produced cylindrical or ellipsoidal Alpha conidia with two oil drops. Pathogenicity tests revealed that both species were pathogenic to fruit of C. sinensis. To our knowledge, the two species were firstly reported on Citrus sinensis in China.
Citrus (Rutaceae) is worldwide cultivated because of its nutritional values and the medicinal benefits (e.g., anti-hypertensive) [Citation1]. In 2018, citrus production in Zigui county, Hubei province, the larger Citrus sinensis production area in China, has reached 23.3 thousand ha. As citrus has diversified as a commercial crop, it became a host for various pathogens from nursery to the storage stage.
Diaporthe species are present as pathogens, endophytes, or saprophytes on a wide range of cultivated or wild trees and ornamentals [Citation2–5]. Species in this genus have been reported as the pathogens of blight, canker, decay, dieback, wilt, leaf spot, fruit rot and root rot across a diverse range of plant species [Citation6–9]. Members of Diaporthe are also frequently associated with citrus diseases worldwide [Citation10,Citation11]. Diaporthe citri (anamorph=Phomopsis citri) caused melanose and stem-end rot of fruit, which are important in most citrus-growing areas with high humidity. Besides, D. foeniculina has also been found from New Zealand, Spain and USA associated with stem end rot on fruit [Citation11]. In China, Diaporthe citri, D. eres and D. unshiuensis have been reported on fruit of Citrus spp. [Citation12].
Preliminarily, species in Diaporthe were identified mainly based on morphological characters and host associations. However, morphology has been conferred to be inconsistent for identification due to inter- and intra-species variability [Citation13]. Molecular analyses inferred that Diaporthe species are not highly host-specific [Citation14]. More than one species is often present on one host, or one species may occur on more than one host [Citation15]. Studies converging on the diversity of Diaporthe have been progressed in recent years in China. Huang et al. (2015) studied Diaporthe on Citrus in China found eight known species and seven novel species based on morphological comparison and multi-gene analyses. Moreover, Diaporthe associated with peach trees [Citation16], pear shoot canker [Citation5], and dieback diseases involving 16 host genera [Citation17] were reported in China. These references provided bounteous information for the study of Diaporthe in China.
During the investigation of fungal pathogens associated with Citrus sinensis in Zigui county, Diaporthe isolates were encountered based on morphology. Three Diaporthe-like isolates, YZU 181047, YZU 181403, and YZU 181223 were found pathogenic to fruit of C. sinenesis. The main objectives of this study were to identify them based on morphological observations and sequence analyses of multiple gene regions.
In 2018, diseased citrus fruit was collected from commercial orchards in Zigui county. Tissues from the margin of infected lesions were cut into segments, which contained both diseased and healthy parts. All segments were surface sterilized in 2% sodium hypochlorite for 2 min, followed by 75% ethanol for 30 s and rinsed in sterile distilled water for three times. All samples were dried with sterile filter paper and plated onto potato dextrose agar (PDA, Difco, USA). Plates were incubated at 25 °C in darkness until mycelia grow. Then, mycelia from colony margin were taken and transferred on fresh PDA plates. Pure cultures were stored in the Fungi Herbarium of Yangtze University (YZU) in Jingzhou, China.
Genomic DNA was extracted from mycelium developed on PDA medium according to Cenis [Citation18]. The primers ITS4 and ITS5 [Citation19] was used to amplify the ITS region of the nuclear ribosomal RNA operon, including the 3′ end of the 18S rRNA, the first internal transcribed spacer region, the 5.8S rRNA gene; the second internal transcribed spacer region and the 5′ end of the 28S rRNA gene. The primers EF1-728F and EF1-986R [Citation20] were used to amplify part of the translation elongation factor 1-α (tef1) gene, and the primers Bt2a and Bt2b [Citation21] were used to amplify the partial beta-tublin (tub2) gene. The PCRs were performed in a 25 μL reaction mixture consisted of 12.5 μL of 2 × Taq PCR StarMix (Genstar, Beijing, China), 2 μL genomic DNA, 1.25 μL of each primer, and 8 μL distilled water (ddH2O). The thermal cycling program was completed on a thermal cycler using the following conditions: initial denaturation at 94 °C for 2 min, followed by 35 cycles of denaturation at 94 °C for 60 s, annealing at (52 °C for ITS, and 56 °C for tef1, and 60 °C for tub2) for 30 s, extension at 72 °C for 60 s, with a final extension step at 72 °C for 5 min. Successful PCR amplification products were purified and sequenced at BGI (Beijing Genomics Institute).
All the obtained sequences were analyzed in the basic-local-alignment search tool (BLASTn) (http://blast.ncbi.nlm.nih.gov/) to retrieve the most similar taxa sequences. Relevant sequences were selected from the studies of Gomes et al. [Citation3], Dissanayake et al. [Citation16], Yang et al. [Citation17], and Tibpromma et al. [Citation22]. All sequences were aligned and combined in the MEGA 7.0 program [Citation23]. Maximum Parsimony (MP) analysis was performed in PAUP version 4.0 b10 [Citation24], generating a heuristic search option of 1000 random-addition replicates and a tree bisection-reconnection (TBR) as a branch-swapping algorithm. MaxTrees were set to 1000, branches of zero length collapsed, and all equally parsimonious trees were saved. Other scores in parsimony were calculated as tree length (TL), consistency index (CI), retention index (RI), and rescaled consistency (RC). The maximum likelihood (ML) [Citation25] phylogeny of the combined dataset was constructed with 1000 bootstrap replicates using GTRGAMMAI model. Additionally, Bayesian (BI) analysis was conducted in MrBayes v. 3.2.6 with 1,000,000 Markov chain Monte Carlo (MCMC) generations and a sampling frequency of every 100th generations. The best-fit evolutionary model was determined via MrModelTest v. 2.3 [Citation26]. At the end of the analysis, the first 25% of the samples were excluded as burn-in, and consensus trees were generated using the 50% majority-rule consensus tree criteria. The tree was viewed in Figtree v.1.3.1 [Citation27]. Posterior probability (PP) values of BI analysis and bootstrap (BP) values of ML and MP analyses were shown at the nodes of branches. The out-group of the phylogeny was Diaporthella corylina CBS 121124.
Three isolates were characterized for their colonial and conidial morphology. Agar plugs (6 mm diam.) of each isolate were taken from the edge of actively growing cultures and transferred onto the center of petri dishes (9 cm diam), containing potato dextrose agar (PDA) and oatmeal agar (OA) for cultural feature. Plates containing 2% water agar (WA) with autoclaved Citrus sinensis leave tissues were incubated at 25 °C under a 12-h near-ultraviolet light/12-h dark cycle to induce sporulation. The culture was checked periodically for the development of ascomata and conidiomata. Morphology was recorded including colony color, texture, microconidia, and sporocarps formation. Conidia were mounted in sterile water for microscopic observation using a light microscope (Nikon DS-Ri2, Tokyo, Japan), equipped with a Nikon DS-Ri2 digital camera. Conidia (n = 50) were measured for each species.
Pathogenicity tests were performed on fruit of Citrus sinensis. Mature and healthy fruit were surface-sterilized in 2% sodium hypochlorite for 2 min and washed three times with sterile distilled water. Fruit was wounded with a sterile scalpel around 6 mm × 6 mm in size. The mycelial plugs from 3 days old cultures grown on PDA were transferred onto the wounds. Controls were treated with sterile PDA. The inoculated fruits were maintained at 25 °C and 80 to 100% relative humidity (RH). The development of disease symptoms was checked daily for one week. The pathogen was re-isolated from the inoculated fruit and identified based on morphology to satisfy Koch's postulates. The pathogenicity tests were conducted with three replicates for each isolate and repeated three times.
A total of nine new sequences were generated and deposited in GenBank (). The combined multi-gene phylogeny (ITS, tef1, and tub2) contained 30 strains, of which 27 were obtained from NCBI (https://www.ncbi.nlm.nih.gov/) (, ). A total of 1276 characters (ITS 494, tef1 377, tub2 405) were included after alignment. Among them, 804 were constant, 190 were variable, and 282 were parsimony uninformative. The heuristic search generated 5 parsimonious trees (TL = 908, CI = 0.664, HI = 0.336, RI = 0.819, RC = 0.544). For the BI analysis, the HKY + I model was recommended by MrModeltest. The topology of ML phylogeny was identical to the results of BI and MP analyses, and it was used as a basal tree.
Figure 1. Phylogram of Diaporthe strains based on combined gene sequences of ITS, tef1 and tub2. Values at the branch nodes indicated maximum parsimony bootstrap (MP BP ≥ 60%), Bayesian posterior (BI PP ≥ 0.6) and maximum likelihood bootstrap (ML BP ≥ 60%), respectively. The tree is rooted with Diaporthella corylina. Strains in the current study are in bold.
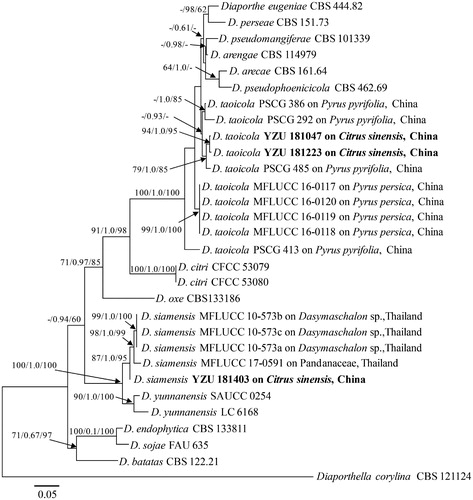
Table 1. Isolates and GenBank accession numbers used in the phylogenetic analyses of Diaporthe.
The phylogenetic tree showed that isolates YZU 181047 and YZU 181223 fell into a clade containing reference strains of Diaporthe taoicola supported with PP values of 0.93 and clustered together with D. taoicola PSCG 485 [Citation5] with the BP or PP values of 94/1.0/95 (MP/BI/ML). The result indicated that both isolates were D. taoicola. However, the isolate YZU 181403 clustered with reference strains of D. siamensis with high BP or PP values of 87/1.0/95 (MP/BI/ML). The result showed that it was D. siamensis.
Morphological examination confirmed the phylogenetic results, the isolates YZU 181047 and YZU 181223 were identified as Diaporthe taoicola [Citation5,Citation16], and the isolate YZU 181403 was D. siamensis [Citation22,Citation28] based on the colony and conidia characteristics.
Diaporthe taoicola Dissanayake, X.H. Li & K.D. Hyde., Mycosphere 8: 543. (2017) ()
Figure 2. Diaporthe taoicola (YZU 181047). (a, b). Front and back view, respectively of colonies on PDA (a) and OA (b); (c). Pathogenicity test on Citrus sinensis fruit for 7 d; (d, e). Conidiomata; (f): Section view of conidiomata; (g). Conidiophores; (h): Alpha conidia. Scale bars: d, e, f = 100μm; g, h: 10 μm.
Sexual morph: Not observed. Asexual morph: Conidiomata 160–230 μm in size, pycnidial, subcuticular, scattered to confluent, dark brown to black, uniloculate, broadly spherical to flattened, cream conidial droplets exuding from central ostioles (). Conidiophores 16–28 × 2–3 μm, hyaline, smooth, densely aggregated, cylindrical, straight, or slightly curved, tapering toward the apex (). Alpha conidia 6–9 × 2–3 μm (av. 8 × 2.7 μm) hyaline, smooth, fusiform to ellipsoid, tapering toward both ends, straight ().
Colony morphology: Colonies on PDA covering the entire Petri dishes after 7 days, ropey with abundant tufted white aerial mycelium, reverse buff with zonate and irregular lines (), 79–81 mm in diam., with aerial mycelium dense in the center and sparse at the marginal area. Colonies on OA flat with white felty aerial mycelium, turning white to dark brown aerial mycelium, conidiomata irregularly distributed on the medium surface after 15-day incubation ().
Materials examined. China, Hubei province, Zigui county, on fruit of Citrus sinensis, August, 2018, M. J. Cui (cultures YZU 181047 and YZU 181223).
Notes: The size and shape of Alpha conidia from the present isolates were identical to that firstly reported by Dissanayake et al. (7–9 × 2–3 μm) [Citation16]. However, Beta conidia were not observed during the present study, which was same as the strains found on pear shoot canker by Guo et al. [Citation5].
Diaporthe siamensis Udayanga, X.Z. Liu & K.D. Hyde., Cryptogamie Mycologie, 33(3): 295-309 (2012) ()
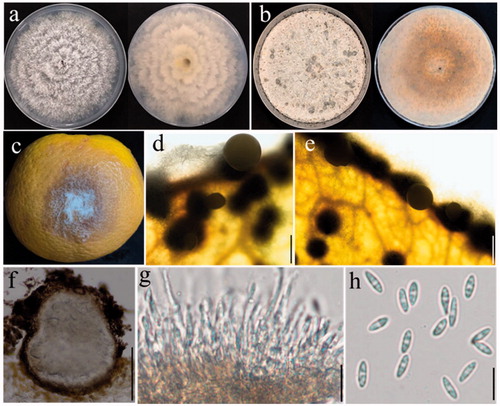
Figure 3. Diaporthe siamensis (YZU 181403). (a, b). Front and back view, respectively of colonies on PDA (a) and OA (b); (c). Pathogenicity test on Citrus sinensis fruit for 7 d; (d, e). Conidiomata; (f): Section view of conidiomata; (g). Conidiophores; (h): Alpha conidia. Scale bars: d, e, f = 100μm; g, h: 10 μm.
Sexual morph: Not observed. Asexual morph: Conidiomata 130–240 μm wide, 94–200 μm high, solitary, single conical neck erumpent through leave tissues, 80–160 × 54–85 μm in size (). Conidiophores 11–23 × 1–2.5 μm, cylindrical, hyaline, straight, or curved, tapering toward the apex (). Alpha conidia 6–8 × 3–3.5 μm (av. 7 × 3.2 μm) hyaline, aseptate, ellipsoidal to oval, biguttulate, rounded at both ends ().
Colony morphology: Colonies on PDA mycelia growing full of Petri-dishes after 7 d with zones of the dirty white and umber, reverse umber patches (). Colonies on OA flat with white felty aerial mycelium, turning white to reddish-brown, with irregular black zones ().
Materials examined. China, Hubei province, Zigui county, on fruit of Citrus sinensis, August, 2018, M. J. Cui (culture YZU 181403).
Note: Alpha conidia of the present isolate were identical to that firstly reported by Udayanga et al. with (3.5–)4–5(–6) × (2–)2.5(–3) μm in size, collected from diseased leaves of Dasymaschalon sp. (Annonaceae) [Citation28]. Besides, its cultural characteristics on PDA were identical to D. siamensis reported by Tibpromma et al., as an endophytic fungus from a Pandanaceae host (Pandanus sp.) [Citation22]. Unfortunately, Beta conidia and Gamma conidia were not observed in the present study.
In the pathogenicity tests, all isolates caused brown fruit rot () on Citrus sinensis, exposing mycelia on surface and severe rotting inside. The initial symptoms appeared as tiny, watery lesions, which gradually expanded eventually led to fruit rot on 7th day. However, the diameters of the lesions varied among different species; D. siamensis caused larger lesions (33–37 mm, av. 34 mm) than D. taoicola (26–30 mm, av. 28 mm) during the tests. In parallel, no lesions developed on the fruit that were inoculated with PDA disks as control. These results showed that all the present isolates were responsible agents for fruit rot of Citrus sinensis.
Presently, the identification of Diaporthe is mainly based on morphological characters and phylogenetic analysis [Citation3,Citation28]. In recent reported studies, nearly 65 Diaporthe species were associated with Chinese hosts, from which 15 were founded on Citrus spp. (). According to Huang et al. [Citation15] and Li et al. [Citation36], phylogeny inferred from combined gene loci of ITS, tef1, and tub2 could be used for further identification of Diaporthe species.
Table 2. Diaporthe species isolated from various hosts in China.
Diaporthe taoicola was firstly isolated from diseased shoots of Prunus persica in Hubei province, China, 2017, proved being able to cause necrotic lesions on detached peach shoots [Citation16]. From then on, it had been only reported on Pyrus pyrifolia causing shoot canker symptoms in China with high phylogenetic diversity [Citation5]. Guo et al. [Citation5] inoculated D. taoicola from pear shoots on wounded twigs of different fruit crops to evaluate its host range, which could induce symptoms on citrus, apple, peach, and kiwifruit. It is worth noting that D. taoicola might pose threats to fruit trees in China. The present study firstly confirmed that it also pathogenic to Citrus sinensis fruit.
Diaporthe siamensis had been reported on diseased leaves of Dasymaschalon sp. in the family of Annonaceae in Thailand [Citation28], but without pathogenicity test on the host plant. Then, it was also found as an endophytic fungus from Pandanus sp. (Pandanaceae) in Thailand [Citation22], also probably as endophyte on Garcinia parvifolia from Malaysia [Citation28]. Regretfully, the pathogenicity evaluation of the species remained a lack in previous studies. Except a detailed description of D. siamensis given in this study, the pathogenicity tests revealed that it could induce fruit rot on Citrus sinensis, stronger than D. taoicola. To the best of our knowledge, this is the first report of Diaporthe taoicola and D. siamensis from Citrus sinensis in China, which could induce fruit rot on the host.
Compliance with ethical standards
The research does not contain any studies with Human Participants and/or Animals, and the authors declare that they have no conflict of interest.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Rauf A, Uddin G, Ali J. Phytochemical analysis and radical scavenging profile of juices of Citrus sinensis, Citrus anrantifolia, and Citrus limonum. Org Med Chem Lett. 2014;4(1):5.
- Santos J, Phillips A. Resolving the complex of Diaporthe (Phomopsis) species occurring on Foeniculum vulgare in Portugal. Fungal Divers. 2009;34:111–125.
- Gomes RR, Glienke C, Videira SIR, et al. Diaporthe: a genus of endophytic, saprobic and plant pathogenic fungi. Persoonia. 2013;31:1–41.
- Gao YHui, Su Y, Sun W, et al. Diaporthe species occurring on Lithocarpus glabra in China, with descriptions of five new species. Fungal Biol. 2015;119(5):295–6146.
- Guo YS, Crous PW, Bai Q, et al. High diversity of Diaporthe species associated with pear shoot canker in China. Persoonia. 2020;45(1):132–162.
- Mostert L, Kang JC, Crous PW. Phomopsis saccharata sp. nov., causing a canker and die-back disease of Protea repens in South Africa. Sydowia. 2011;53:227–235.
- Udayanga D, Liu X, McKenzie EHC, et al. The genus Phomopsis: biology, applications, species concepts and names of common phytopathogens. Fungal Diversity. 2011;50(1):189–225.
- Du ZHUO, Fan X-L, Hyde KD, et al. Phylogeny and morphology reveal two new species of Diaporthe from Betula spp. in China. Phytotaxa. 2016;269(2):90–102.
- Díaz GA, Latorre BA, Lolas M, et al. Identification and characterization of Diaporthe ambigua, D. australafricana, D. novem, and D. rudis causing a postharvest fruit rot in kiwifruit. Plant Dis. 2017;101(8):1402–1410.
- Timmer LW, Garney SM, Graham J. Compendium of citrus diseases. 2nd ed. St. Paul, Minnesota, USA: APS Press; 2000. 19–21.
- Udayanga D, Castlebury LA, Rossman AY, et al. Species limits in Diaporthe: molecular re-assessment of D. citri, D. cytosporella, D. foeniculina and D. rudis. Persoonia. 2014;32:83–101.
- Huang F, Udayanga D, Wang XH, et al. Endophytic Diaporthe associated with citrus: a phylogenetic reassessment with seven new species from china. Fungal Biol. 2015;119(5):331–347.
- Santos JM, Correia VG, Phillips AJL. Primers for mating-type diagnosis in Diaporthe and Phomopsis: their use in teleomorph induction in vitro and biological species definition. Fungal Biol. 2010;114(2-3):255–270.
- Rehner SA, Uecker FA. Nuclear ribosomal internal transcribed spacer phylogeny and host diversity in the coelomycete Phomopsis. Can J Bot. 1994;72(11):1666–1674.
- Huang F, Hou X, Dewdney M, et al. Diaporthe species occurring on citrus in China. Fungal Diversity. 2013;61(1):237–250.
- Dissanayake AJ, Zhang W, Liu M, et al. Diaporthe species associated with peach tree dieback in Hubei. China. Mycosphere. 2017;8(5):533–549.
- Yang Q, Fan XL, Vladimiro G, et al. High diversity of Diaporthe species associated with dieback diseases in China, with twelve new species described. MC. 2018;39:97–149.
- Cenis JL. Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids Res. 1992;20(9):2380.
- White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, et al., editors. PCR protocols: a guide to methods and applica-tions., San Diego, California: Academic Press; 1990. 315–322.
- Carbone I, Kohn LM. Method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91(3):553–556.
- Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Am Soc Microbiol. 1995;61:1323–1330.
- Tibpromma S, Hyde KD, Bhat JD, et al. Identification of endophytic fungi from leaves of Pandanaceae based on their morphotypes and DNA sequence data from southern. MycoKeys. 2018;33:25–67.
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Evolutionary Gene-tics. Analysis. 2016;33:1870–1874.
- Swofford DL. Phylogenetic analysis using parsimony. Version 4b10. Sunderland, MA: Sinauer Associates; 2002.
- Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948.
- ThailNylander JAA. MrModelTest v. 2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University. 2004.
- Rambaut A, Drummond A. FigTree v.1.3.1. Institute of Evolutionary Biology. Edinburgh, UK: University of Edinburgh; 2010.
- Udayanga D, Liua X, Mckenzie EHC, et al. Multi-locus phylogeny reveals three new species of Diaporthe from Thailand. Cryptogamie Mycologie. 2012; 33(3):295–309.
- Gao YH, Liu F, Duan WJ, et al. Diaporthe is paraphyletic. Ima Fungus. 2017;8(1):153–187.
- Bai Q, Zhai LF, Chen XR, et al. Biological and molecular characterization of five Phompsis species associated with pear shoot canker in China. Plant Disease. 2015;99(12):1704–1712.
- Hu DM, Cai L, Hyde KD. Three new ascomycetes from freshwater in China. Mycologia. 2012;104(6):1478–1489.
- Yang Q, Fan XL, Du Z. Diaporthe juglandicola sp. nov. (Diaporthales, Ascomycetes), evidenced by morphological characters and phylogenetic analysis. Mycosphere. 2017;8(5):817–826.
- Crous PW, Shivas RG, Quaedvlieg W, et al. Fungal planet description sheets: 214. Persoonia. 2014;32(1):184–280.
- Fan XL, Hyde KD, Udayanga D, et al. Diaporthe rostrata, a novel ascomycete from Juglans mandshurica associated with walnut dieback. Mycol Progress. 2015;14(10):82.
- Yang Q, Du Z, Tian CM. Phylogeny and morphology reveal two new species of Diaporthe from Traditional Chinese Medicine in Northeast China. Phytotaxa. 2018;336(2):159–170.
- Li Y, Tan P, Zhao DG. Diaporthe nobilis, a new record on Camellia sinensis in Guizhou Province. China. Mycosphere. 2017;8(1):1–8.
