Abstract
A fungus of the genus Kordyana, found on leaves of Commelina communis and C. minor exhibiting white smut-like symptoms, was identified as Kordyana commelinae based on morphological characteristics and two rDNA sequence analyses. We report the novel occurrence of the genus Kordyana in Korea and the association of K. commelinae with the host plant species. As well, we provide the necessary mycological information to resolve species delimitation and taxonomic problems of Kordyana.
Commelina communis L. (Commelinaceae) is an annual monocot native throughout East Asia, including China, Japan, Korea, Laos, Russia (Far East), Thailand, and Vietnam [Citation1]. Within its native range, C. communis is a common forest groundcover species with attractive blue flowers. Conversely, C. communis has become an aggressive invader of natural ecosystems and crop fields in the United States and the Czech Republic, where it has been introduced as an ornamental species [Citation2]. The species is known to be challenging to control owing to its aggressive growth, seed longevity, broad ecological tolerance of climatic and soil factors, and glyphosate tolerance [Citation2]. Commelina minor, which was newly found in Korea in 1981, appears to be naturally distributed across the country in localized populations [Citation3]. This species is similar in appearance to C. communis except having smaller leaves and pale pink flowers [Citation3].
Fungi in Kordyana, Brachybasidiaceae (Exobasidiales, Basidiomycota), are leaf pathogens causing a white smut-like disease, typically on members of Commelinaceae [Citation4]. Several species of Kordyana, in particular K. brasiliensis, K. tradescantiae, and K. celebensis, are considered promising candidates for biocontrol of invasive weeds, such as Tradescantia fluminensis in Australia, New Zealand, and China and C. benghalensis in the United States and China [Citation4–6]. Although Kordyana is the oldest genus in Brachybasidiaceae, knowledge at the genus and species level is still incomplete owing to taxonomic problems and the lack of morphological descriptions and DNA sequencing for some species [Citation7]. In this study, we identified and described a species of Kordyana, newly found on C. communis and C. minor, based on morphological characteristics and molecular phylogenetic analyses.
Leaves of C. communis and C. minor displaying white smut-like symptoms were collected in Wonju, Jeonju, Seoul, and Hongcheon, Korea, from August through September 2020. Voucher specimens were deposited in the Korea University herbarium (KUS –F31903 and F31925 for C. communis; F31911 and F31953 for C. minor). Fungal isolation was achieved by fixing small pieces of leaf with newly sporulating lesions to the inside of the lid of a sterile Petri dish filled with 2% Potato Dextrose Agar (PDA, Difco, Rockville, MD). The lid was then turned at 2-h intervals. Individual colonies that developed on the agar were sub-cultured onto new PDA plates and incubated in the dark at 25 °C for 2 weeks. Representative isolates for each specimen were deposited in the Korean Agricultural Culture Collection (accession nos. KACC49694, 49695, 49696, and 49697). Using fresh plants, we prepared glass slide specimens of the abaxial leaf epidermis (including tufts of basidia and basidiospores) stained with lactophenol cotton blue. The mounted specimens were viewed under compound light microscopes (Olympus BX51, Olympus, Tokyo, Japan; Zeiss AX10 microscope equipped with an AxioCam MRc5, Carl Zeiss, Jena, Germany).
To confirm the morphological identification, genomic DNA was extracted from four cultures, two for each of the two host species, using Quick-DNA Fungal/Bacterial Miniprep Kit (Zymo Research, Irvine, CA). The internal transcribed spacer (ITS) and large-subunit (LSU) regions of the fungal rDNA were amplified [Citation8] and sequenced using the primer pairs ITS1/ITS4 [Citation9] and LR0R/LR5 [Citation10], respectively. The obtained sequences were compared to those in GenBank and the Genetic Resource Center (GRC), National Agriculture and Food Research Organization (NARO) of Japan (https://www.gene.affrc.go.jp) using a BLAST search. For phylogenetic analyses, both ITS and LSU sequences from all Exobasidiales genera in GenBank and the GRC were selected. Rhamphospora nymphaeae (GenBank accession nos. NR119615 and NG057757) was used as an outgroup taxon per the phylogenetic results of Piepenbring et al. [Citation7]. Two phylogenetic trees were constructed for the sequences of both the ITS and LSU regions using MEGA7 software [Citation11] based on neighbor-joining (NJ) analysis with the following conditions: gaps in the sequence alignment were treated as deletions, evolutionary distances were calculated using the Kimura-2-parameter model, including substitutions of transitions and transversions, and bootstrap values for branches were calculated using one thousand replicates.
Flat lesions on the upper leaf surface initially appeared as diffuse chlorotic spots. As chlorosis spread, the lesions became more prominent, circular in outline, and in some instances, confluent without a conspicuous margin. Lesions on the lower leaf surface were initially discrete, circular, and whitish in color, turning yellow to dark-brown from the center of the lesion as necrosis progressed (). All substomatal chambers within abaxial lesions were filled with fungal hyphae and whitish caespituli (tufts of basidia and basidiospores) protruded directly from stomatal openings with a dense and even distribution (). The presence of balls of basidia developing through stomatal openings is one of distinctive characteristics of the genus Kordyana [Citation7].
Figure 1. White smut-like disease of Commelina communis and C. minor associated with Kordyana commelinae. (a) Infected leaves with flat lesions in the field. (b) Abaxial side of lesions consisting of whitish caespituli. (c) Close-up of whitish caespituli. (d) Basidia emerging from a stomatal opening. (e) A basidium with two basidiospores. (f) A basidiospore. (g) A basidiospore producing conidia on germ tubes. (h) Conidia. (i) A budding conidium. (j) A gelatinous, corrugate, and cream-colored colony on PDA (3-week-old). (a)–(c), (f), (h), and (i) from C. minor. (d), (e), (g), and (j) from C. communis.

Kordyana commelinae was consistently isolated from the symptomatic leaf tissues. Basidia were clavate, hyaline, and bisterigmatic, measured 18–34 × 4 μm (avg. 25.9 × 4 μm, n = 20), and were directly emergent from stomatal openings (). Basidiospores were clavate to reniform, hyaline, and measured 7–13 × 2–4 μm (avg. 10.2 × 2.3 μm, n = 30) (). Conidia were linear, acicular, and measured 2–6.5 × 0.5–1.5 μm (avg. 3.2 × 1.1 μm, n = 30) (). Conidia were produced laterally from–or at the tip of–a basidiospore germ tube, and from conidia by budding (). When cultured on PDA, colonies were slow-growing, gelatinous, corrugate, and cream to pale yellow (). The morphological characteristics of basidia and basidiospores were most consistent with those of K. commelinae as originally described on C. nudifera by Petch [Citation12] and distinct from the other Kordyana species. The basidia of the fungus we report here were shorter than those of K. brasiliensis (66–76 μm) and K. celebensis (up to 60 μm) [Citation4,Citation13]. The basidiospores of the fungus were shorter than those of K. tradescantiae (15–18 μm) and K. polliae (15–21 μm), but more slender than those of K. pinangae (6–11 × 4–6 μm) [Citation4]. Diagnostic descriptions of K. indica are lacking, and it is doubtful that the fungus is any of the other three species (i.e., K. boswelliae, K. cyphelloidis, and K. polliniae) given that their hosts are not in the family Commelinaceae [Citation4,Citation7].
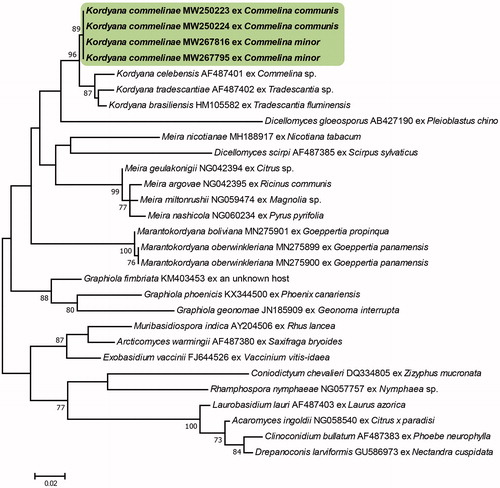
The ITS sequences of our four Korean isolates had 97.47–100% similarity to that of a K. commelinae isolate (MAFF239954) in the GRC database. Unfortunately, there were no ITS sequences available for Kordyana spp. in the GenBank database. The isolates obtained from C. communis and C. minor were distinguished based on their ITS sequences, which differed by 12 base pairs, including 10 gaps. The LSU sequences of the Korean isolates were identical to each other and showed the most similarity (94.77–95.85%) to Kordyana spp. (MH908997, KX348017, AF487401, AF487402, KX348016, and MH909025). It is probable that K. commelinae might have genetic differences at the sub-species level, depending on the host plant species, but in-depth studies with more isolates obtained from diverse Commelina spp. needed to address the issue. The NJ analysis of the sequences of the ITS and LSU regions confirmed that the Korean specimens are in the same clade as the K. commelinae strain obtained in Japan and different from the other Kordyana species and Exobasidiales genera ( and ).
Figure 2. Phylogenetic relationship of Kordyana commelinae specimens and reference sequences retrieved from GenBank and the Genetic Resource Center, National Agriculture and Food Research Organization of Japan, inferred from neighbor-joining analysis using the ITS sequences. Bootstrap values (1000 replicates) above 70% are indicated at the branches. The Korean specimens are indicated in bold. The scale bar represents 0.05 nucleotide substitutions per site.
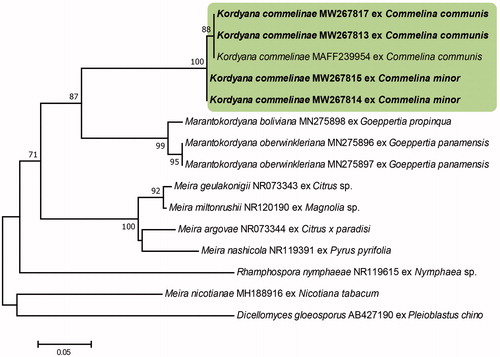
Figure 3. Phylogenetic relationship of Kordyana commelinae specimens and reference sequences retrieved from GenBank, inferred from neighbor-joining analysis using the LSU sequences. Bootstrap values (1000 replicates) above 70% are indicated at the branches. The Korean specimens are indicated in bold. The scale bar represents 0.02 nucleotide substitutions per site.
This is the first collection of the genus Kordyana in Korea, and the first description of the presence of K. commelinae on C. communis and C. minor. The fungus was initially found on leaves of C. nudiflora in Sri Lanka, and its morphological characteristics were reported by Petch as indicative of a new species [Citation12]. Although there are two more records of K. commelinae from China and Taiwan [Citation6], no details on symptomatic and morphological features were provided in these reports. A record of culture collection of the species and related DNA sequence data were found in the GRC, NARO of Japan, but there have been no reports concerning the finding in Japan. With the present study, we have provided additional information on symptomatic, morphological, and cultural characteristics, culture collections, and data for two gene sequences of K. commelinae found on two Commelina species in Korea.
Commelina communis is an invasive and noxious weed found in gardens and crop fields in North America and the Czech Republic and can also cause serious damage in orchards and tree plantations within its native range in China and Japan [Citation2]. The species is reported to be challenging to control in glyphosate tolerant crop systems [Citation2]. Kordyana species have been studied and are considered promising biocontrol agents of the highly invasive weeds in Commelinaceae [Citation4]. This novel collection of K. commelinae in Korea, along with the information we have provided, is expected to be a valuable source for further studies on the biological control of C. communis. As well, we hope that the study contributes to further understanding of Kordyana species, especially with regard to species delimitation and taxonomic problems [Citation7].
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- eFloras. Published on the internet http://www.efloras.org. [accessed 21 October 2020]. St. Louis, MO: Missouri Botanical Garden; Cambridge, MA: Harvard University Herbaria; 2008.
- Gomez JM. Glyphosate-tolerant Asiatic dayflower (Commelina communis L.): ecological, biological and physiological factors contributing to its adaptation to Iowa agronomic systems [master thesis]. Ames, IA: Iowa State University; 2012.
- Lee YN. New taxa of Korean flora (3). Korean J Bot. 1981;24:27–30.
- Macedo DM, Pereira OL, Hora Júnior BT, et al. Mycobiota of the weed Tradescantia fluminensis in its native range in Brazil with particular reference to classical biological control. Australas Plant Pathol. 2016;45(1):45–56.
- Ellison CA, Barreto RW. Prospects for the management of invasive alien weeds using co-evolved fungal pathogens: a Latin American perspective. Biol Invasions. 2004;6(1):23–45.
- Farr DF, Rossman AY. Fungal Databases, U.S. National Fungus Collections, ARS, USDA. Retrieved December 2, 2020, from https://nt.ars-grin.gov/fungaldatabases/.
- Piepenbring M, Hartmann M, Hofmann TA, et al. Two new species in a new genus and a critical revision of Brachybasidiaceae (Exobasidiales, Basidiomycota) in honor of Franz Oberwinkler. Mycol Prog. 2020;19(4):351–365.
- White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TH, editors. PCR protocols: a guide to methods and applications. San Diego, CA: Academic Press; 1990. p. 315–322.
- Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes – application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2(2):113–118.
- Moncalvo JM, Lutzoni FM, Rehner SA, et al. Phylogenetic relationships of agaric fungi based on nuclear large subunit ribosomal DNA sequences. Syst Biol. 2000;49(2):278–305.
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874.
- Petch T. Additions to Ceylon fungi (II). Ann Roy Bot Gard (Peradeniya). 1922;7:279–320.
- Gäumann E. Über die Gattung Kordyana Rac. Ann Mycol. 1922;20:257–271.
