Abstract
During the rainy season in Thailand, specimens of Phallus chiangmaiensis sp. nov. and P. merulinus were collected from Chiang Mai and Samut Sakhon Provinces, respectively. Molecular phylogenetic analyses based on sequences of the nuclear ribosomal large subunit (LSU), nuclear ribosomal 5.8S gene including the internal transcribed spacer regions 1 and 2 (ITS), and the protein-coding gene atp6 (mitochondrial adenosine triphosphate [ATP] synthase subunit 6) support the placement of the new species within Phallus. Phallus chiangmaiensis has a well-developed white indusium and campanulated caps with reticulate surfaces. It differs morphologically from the related species, as supported by the phylogenetic data. Phallus merulinus is reported here as a species that was re-encountered in Thailand. The descriptions of the species are accompanied by illustrations of macro- and micro- morphological features, and a discussion of the related taxa is presented.
Keywords:
Introduction
Species in the genus Phallus Junius ex L., commonly known as stinkhorn, are gasteriod fungi in the family Phallaceae, order Phallales, with P. impudicus L. as the type species. The genus is characterized by a fetid odor originating from the gleba. The important morphological features used for species delimitation are the shape and surface configuration of the receptacle, the coloration of the receptacle, volva with rhizomorphs, the presence of an erect to curved sponge-like and hollow pseudostipe, the size of the basidiomata, and the presence or absence of indusia (skirt-like structures) [Citation1–3].
Some species of Phallus, including P. atrovolvatus Kreisel & Calonge, P. dongsun T.H. Li, T. Li, Chun Y. et al., P. echinovolvatus (M. Zang & Z.X. Hu) Kreisel, P. fragrans M. Zang, P. fuscoechinovolvatus T.H. Li, B. Song & T. Li, P. impudicus, P. indusiatus Ventenat., P. luteus (Liou & L. Hwang) T. Kasuya, P. merulinus (Berk.) Cooke, P. mengsongensis H.L. Li, L. Ye, P.E. Mortimer et al., P. nanchangensis Z.Z. He, and P. rubrovolvatus (M. Zang, D.G. Ji & X.X. Liu) Kreisel are used for food [Citation4–8]. Phallus rubicundus (Bosc) Fr. and P. tenuis (E. Fisch.) Kuntze are inedible species of Phallus that are used as medicines [Citation6,Citation9].
Currently, Phallus consists of 95 species, excluding formae, varieties and synonyms, according to the Index Fungorum database (www.indexfungorum.org). Phallus is widely distributed in different geographical locations and climate types, such as grasslands, conifer forests, bamboo forests, and broadleaved forests from tropical, subtropical, and temperate areas [Citation3,Citation7,Citation10–17].
During surveys of wild mushrooms in Thailand, we found a new species of Phallus, as supported by morphological and phylogenetic analyses. We introduced this new species to the Phallaceae (Phallales, Agaricomycetes). Another species was identified as Phallus merulinus which has previously been reported from Thailand [Citation18,Citation19].
Materials and methods
Fungal specimen
Two fresh specimens from the Saluangnok community forest, Chiang Mai Province, and twelve fresh specimens from Amphoe Ban Phaeo, Samut Sakhon Province, Thailand were collected during the rainy season of 2019.
Isolation and morphological studies
Photographs of the fresh specimens in their natural habitat were taken from different angles with a digital camera (Canon, EOS 60 D, Canon Marketing Co., Ltd., Bangkok, Thailand) for further studies, and field notes relating to possible host plants and the situations in which the fruit bodies were found were documented. The fresh basidiocarps were wrapped in wax paper and carefully handled to a laboratory for isolation. The macroscopic features used for identification, such as color, size, shape, outer surface of the fruiting body, and ecological and host substrates, were recorded. The colors of the fresh specimens were described using The RHS color chart, a sixth revised edition [Citation20].
The small pieces of endoperidium tissue of the fruiting bodies were aseptically transferred to the potato dextrose agar plates (PDA; Difco, Becton, Dickinson and Company, Bangkok, Thailand) with antibiotics (penicillin G (0.05 g/L) and streptomycin sulfate (0.05 g/L)). The plates were incubated at room temperature (25 °C). The mycelia emerging from the tissue were transferred to the new PDA plates. The specimens were dried by a dehydration machine at 45 °C for 24–36 h and deposited in the BIOTEC Bangkok Herbarium (BBH), Thailand.
The hand section of the dried specimens was made under an Olympus SZ61 (Olympus Co., Ltd., Bangkok, Thailand) and the sections were mounted in 5% KOH solution and 1% Congo Red. Morphological characteristics, such as size, color, and shape of basidiospores; and the cells or hyphae of the cap, pseudostipe, indusium, volva, and rhizomorph, were examined under an Olympus BX31 light microscope. Micrographs were obtained with an Olympus microscope equipped with differential interference contrast (Olympus DP70, Olympus Co., Ltd., Bangkok, Thailand) and a Canon EOS 60 D camera. The growth rate and colony characteristics were recorded from the cultures grown on the PDA. The cultures were deposited in BIOTEC Culture Collection (BCC), Thailand. The fungal taxonomic details were also submitted to Faces of Fungi and Index Fungorum.
DNA extraction and PCR amplification
Genomic DNA was extracted from the mycelia on PDA using a CTAB method [Citation21]. The LSU, ITS, and atp6 gene regions were amplified using the primer pairs LROR/LR5, ITS5/ITS4, and 1M40F/2M, respectively [Citation22–24]. The amplification reactions were performed in a 50 µl reaction volume containing 38.3 µl of ddH2O, 5.0 µl of 10× buffer, 2.5 µl of MgCl2, 1.0 µl of dNTP, 1 µl of each primer (10 µM), 0.2 µl of Taq DNA polymerase (Vivantis, Bang Trading 1992 Co., Ltd., Bangkok, Thailand) and 1 µl of DNA template. The amplification conditions for the LSU and ITS regions followed the protocol described by Sakayaroj [Citation23], while the amplification conditions for the atp6 gene followed the protocol described by Raspé et al. [Citation24]. The PCR products were sequenced using the same primers as used for amplification.
Sequence alignment and phylogenetic analyses
Individual analyses were run for separate loci (ITS dataset consisting of 42 sequences, LSU 34 sequences, atp6 19 sequences) and a combined analysis comprising ITS, LSU, and atp6 (46 sequences) shown in . Sequences were assembled using BioEdit v.7.0.5.3 [Citation25]. All sequences were aligned with MUSCLE [Citation26] and manually edited using BioEdit v.7.0.5.3 [Citation25]. The phylogenetic analyses were performed using maximum likelihood (ML), maximum parsimony (MP), and Bayesian inference (BI).
Table 1. Taxa used in the phylogenetic analyses and the new taxa in bold.
The maximum likelihood analysis was performed on the CIPRES supercomputer using the program RAxML-HPC2 v.8.2.12 on XSEDE [Citation27]. One thousand nonparametric bootstrap iterations were run with the GTR model and a discrete gamma distribution.
The maximum parsimony analysis was performed by PAUP v.4.0b10 [Citation28] with 10 replicates of stepwise additions, the heuristic search option, 1,000 random taxa addition and the tree-bisection reconnection (TBR) branch-swapping algorithm. All characters were given equal weight, and gaps were treated as missing data. Maxtrees were unlimited, branches of zero length were collapsed, and all multiple, equally parsimonious trees were saved. The robustness of the most parsimonious tree was estimated based on 1,000 bootstrap replications.
The Bayesian analysis was performed MrBayes v.3.0b4 [Citation29] using a uniform [GTR + I + G] model, Isetnst = 6 rates = invgamma; prsetstatefreqpr = dirichlet (1,1,1,1). Four Markov chains were run for 5,000,000 generations, and trees were sampled every 100 generations. The first 5,000 trees, which represented the burn-in phase of the analysis, were discarded, with 50,000 trees used for calculating posterior probabilities (BIPP) in the consensus tree.
Results
Phylogenetic analyses
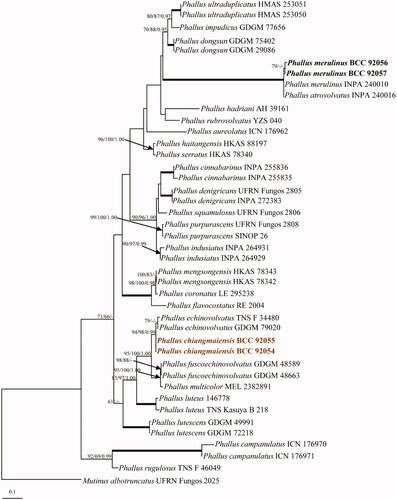
The ITS dataset included 42 sequences, and Mutinus albotruncatus (UFRN Fungos 2025) was used as an outgroup [Citation30]. The best scoring of the RAxML tree is shown in , with the final optimization likelihood value of −5932.961212. The maximum parsimony dataset consists of 789 characters, of which 347 were constant, 102 were variable parsimony-uninformative and 340 were parsimony informative with a length of 1,168 steps (CI = 0.615, RI = 0.783, RC = 0.481 and HI = 0.385). Bootstrap support values for maximum likelihood (BSML, left), maximum parsimony (BSMP, middle) were >60%. Branches with Bayesian posterior probabilities (BPP, right) >0.95 are indicated at the nodes. The two strains of Phallus chiangmaiensis sp. nov. (BCC 92054 and BCC 92055), are closely related to P. echinovolvatus with bootstrap and posterior probability strong support (94% BSML, 98% BSMP and 0.99 BPP), shown in . However, the morphological analyses that our new species and P. echinovolvatus are distinct. Phylogenetic trees generated from LSU and atp6 sequences can be seen in Supplementary Figures 1 and 2.
Figure 1. Phylogenetic relationships of Phallus spp. inferred from ITS sequences. Numbers at the significant nodes represent ML bootstrap values/MP/Bayesian posterior probabilities, multiplied by 100; bold lines in the tree represent 100% bootstrap (BSMP, BSML) and 1.00 posterior probability (BPP).
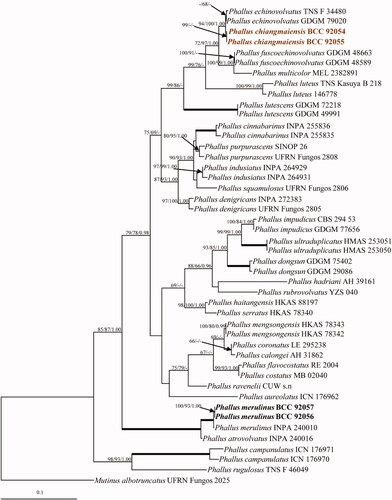
The dataset of combined genes (ITS, LSU, and atp6), Mutinus albotruncatus (UFRN Fungos 2025) was used as an outgroup. The best scoring of the RAxML tree is shown in , with the final optimization likelihood value of −11436.786086. The maximum parsimony dataset consists of 2,438 characters, of which 1,715 were constant, 208 were variable parsimony-uninformative and 515 were parsimony informative with a length of 1,737 steps (CI = 0.625, RI = 0.770, RC = 0.481 and HI = 0.375). Bootstrap support values for maximum likelihood (BSML, left), maximum parsimony (BSMP, middle) were >60%. Branches with Bayesian posterior probabilities (BPP, right) >0.95 are indicated at the nodes. The phylogenetic analyses showed that all the collected strains were clustered in the family Phallaceae. The two strains of Phallus chiangmaiensis sp. nov. (BCC 92054 and BCC 92055), which were recovered as a distinct species, grouped with P. echinovolvatus, P. fuscoechinovolvatus, P. multicolor P. lutescens, and P. luteus and were separated from other species with bootstrap support (99% BSML and 86% BSMP). Both strains of Phallus merulinus (BCC 92056 and BCC 92057) clustered with P. merulinus (INPA 240010), with high statistical support (100% BSMP, 100% BSML, and 1.00 BPP) in the tree ().
Figure 2. Phylogenetic relationships of Phallus spp. from a combined ITS, LSU, and atp6 analyses. Numbers at the significant nodes represent ML bootstrap values/MP/Bayesian posterior probabilities, multiplied by 100; bold lines in the tree represent 100% bootstrap (BSMP, BSML) and 1.00 posterior probability (BPP).
Taxonomy
Phallus chiangmaiensis U. Pinruan, S. Sommai & P. Khamsuntorn, sp. nov.
Figure 3. Phallus chiangmaiensis (BBH 47825, holotype). (a) Mature basidiomata. (b) Reticulate cap. (c) Indusium. (d) Immature basidiomata (egg). (e) Pseudostipe and section of immature basidiomata. Scale bars: a = 50 mm, b–e = 10 mm.

Figure 4. Microscopic features of Phallus chiangmaiensis. (a,b) Cap cells and hyphae. (c) Cells of indusium. (d) Cells of pseudostipe. (e) Crystals in volva hyphae (arrowed). (f,g) Volva hyphae with clamp connections (arrowed). (h,i) Rhizomorph hyphae with clamp connections (arrowed). (j) Basidia with sterigmata and basidiospores. (k) Basidiospores. (l) Colony on PDA (surface and reverse plate). Scale bars: a, c–e = 20 µm, b, f–j = 10 µm, g, k = 5 µm, l = 10 mm.
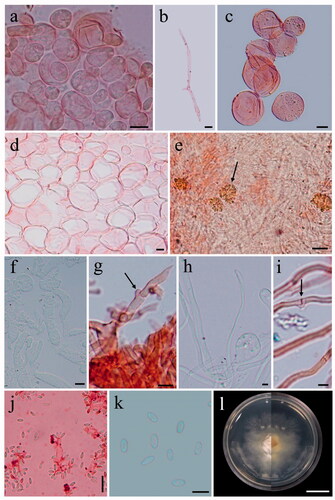
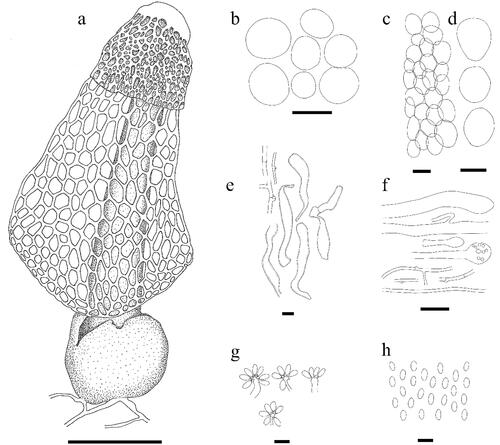
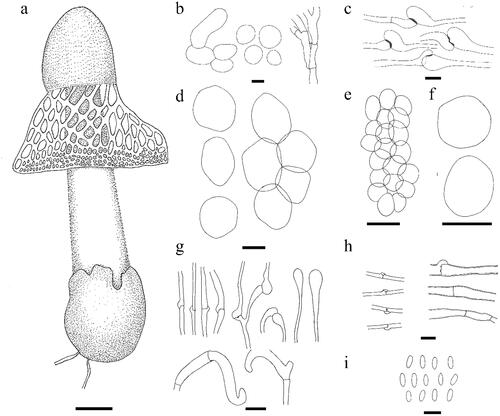
Figure 5. Line drawing of Phallus chiangmaiensis. (a) Fruiting body. (b) Cells of indusium. (c,d) Cells of pseudostipe. (e) Volva hyphae. (f) Rhizomorph hyphae. (g) Basidia. (h) Basidiospores. Scale bars: a = 50 mm, b–d = 50 μm, e = 10 μm, f = 20 μm, g–h = 5 μm.
Index Fungorum number: IF557726; Facesoffungi number: FoF 08402
Etymology: The name refers to Chiang Mai Province, the location where the mushroom was collected.
Asexual morph: Unknown.
Holotype: BBH 47825
Sexual morph: Egg globose to. subglobose, 22–30 mm in diam., white (RHS2015 N155C) with a white mycelial rhizomorph arising from the base. Exoperidium papery, milky white (RHS2015 N155C); mesoperidium gelatinous or lightly viscous, transparent to subtransparent, 3.5–5 mm thick, moderate yellowish-brown (RHS2015 N199C); endoperidium membranous, thin, white (RHS2015 N155C), covering an upper surface of gleba. Mature basidiomata 205–215 mm high. Cap campanulate, 40–50 mm high, 35–45 mm wide, surface strongly reticulate, light yellow (RHS2015 162C), meshes deep, polygonal, apex with an apical pore and covered with greenish-white (RHS2015 155C) membrane approximately ¼ the size of the cap. Gleba moderate olive-brown (RHS2015 199A), mucilaginous. Pseudostipe 158–165 mm high, cylindrical, tapering toward the apex, 20–25 mm wide at the base, 10–13 mm wide at the apex, white (RHS2015 NN155D), fragile and soft, spongy, hollow. Indusium coarsely latticed, white (RHS2015 NN155D), extended to ¾ the size of the pseudostipe. The meshes of indusium are large, hexagonal or polygonal, 5–10 mm wide. Volva globose to subglobose, 55 × 40 mm in diam., light brownish gray (RHS2015 201B), smooth surface, Rhizomorphs white (RHS2015 NN155D), when scratched the color changes to light purple (RHS2015 85B). Odor fetid.
Basidia 7.0–15.0 × 1.5–3.0 μm, elongated, cylindrical, slightly broader at the center, hyaline. Sterigmata 4–8 in number. Basidiospores 3.0–4.0 × 1.5–2.0 μm (x̅ = 3.9 × 1.9 μm, n = 55), ellipsoid, greenish-white (RHS2015 192D) in 5% KOH, inamyloid, smooth surface and thin-walled. Cap cells and hyphae; cells 12.5–25 μm in diam., globose to subglobose, hyaline, thin-walled; hyphae 2.0–10.0 μm wide, hyaline, thin-walled, septate, branched with clamp connections. Cells of pseudostipe 15.0–67.5 μm in diam., pseudoparenchymatous, globose to a subglobose, bubble-like, hyaline, smooth surface and thin-walled. Cells of indusium 12.5–57.5 μm in diam., hyaline, globose to subglobose or bubble-like, smooth surface, thin-walled. Volva hyphae composed of two types of hyphae; type I: 2.0–2.5 μm wide, hyaline, septate, branched, smooth surface, thin-walled with clamp connections, type II: 5.0–10.0 μm wide, irregular shape, hyaline, septate, branched smooth surface and thick-walled; crystal deposits in globose to subglobose cells distributed among the hyphae. Rhizomorph hyphae are composed of two types of hyphae; type I: 2.5–5.0 μm wide, hyaline, septate, branched, smooth surface, thin-walled with clamp connections, type II: 10.0–15.0 μm wide, hyaline, branched, smooth surface, thin-walled, swollen at the tip.
Known distribution: Saluangnok community forest, Amphoe Mae Rim, Chiang Mai Province, Thailand.
Habit and Habitat: Solitary or scattered on soil, under Bambusa sp.
Culture characteristics: Tissue germinated on PDA within 24 h. Colonies were grown on PDA with scant mycelium, entire margin, reaching 2.0 cm in diam. in 1 month at 25 °C, surface and reverse white to cream.
Materials examined: THAILAND, Chiang Mai Province, on soil under Bambusa sp., October 8 2019, U. Pinruan, (holotype BBH 47825, isotype BBH 49056); culture ex-holotype BCC 92054, culture ex-isotype BCC 92055.
Notes: Phylogenetically, Phallus chiangmaiensis is most closely related to P. echinovolvatus. Morphologically, it differs from P. echinovolvatus on the surface of volva. In P. echinovolvatus the volva is echinulate while in P. chiangmaiensis it is smooth. The cap of P. chiangmaiensis is larger (40–50 × 25–45 mm) than of P. echinovolvatus (25–30 × 25–30 mm). The length of indusium in P. chiangmaiensis is longer (130–160 mm) than in P. echinovolvatus (70–100 mm). The basidia of P. echinovolvatus are 6–8 μm long with 4–6 sterigmata while those of P. chiangmaiensis are up to 15 μm long with up to 8 sterigmata. The phylogenetic analyses show that our new species also grouped with P. fuscoechinovolvatus, P. multicolor, P. lutescens and P. luteus. However, it morphologically differs from P. multicolor and P. luteus in having a white indusium, and from P. fuscoechinovolvatus in having non-echinulated volva, as shown in .
Table 2. Synopsis of macro- and micro- characteristics of Phallus chiangmaiensis, P. echinovolvatus, P. fuscoechinovolvatus, P. lutescens, P. luteus, and P. multicolor.
Phallus merulinus (Berk.) Cooke (1882)
Figure 6. Phallus merulinus (BBH 47826). (a,b) Basidiomata. (c,d) Immature basidiomata (eggs) with rhizomorph (arrowed). (e) Indusium and pseudostipe. (f) Volva. Scale bars: a–b = 50 mm, c–f = 20 mm.
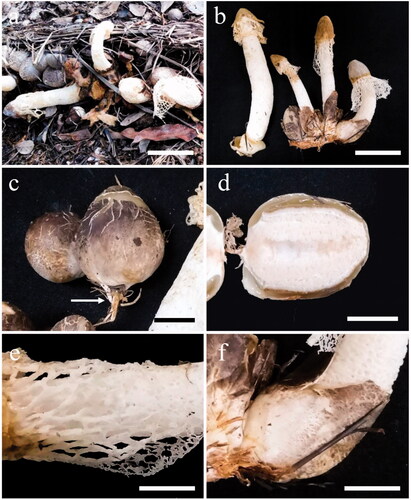
Figure 7. Microscopic features of Phallus merulinus. (a–e) Cap cells and hyphae. (f) Cells of indusium. (g) Cells of pseudostipe. (h,i) Volva hyphae. (j,k) Rhizomorph hyphae. (l) Basidiospores. (m) Colony on PDA (surface and reverse plate). Scale bars: a–b, f–g = 20 µm, c–e, h–l = 5 µm, m = 20 mm.
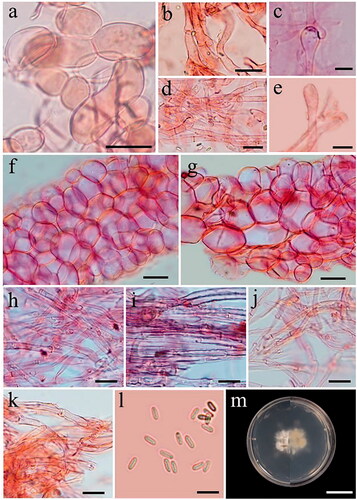
Figure 8. Line drawing of Phallus merulinus. (a) Fruiting body. (b,c) Cap cells and hyphae. (d) Cells of indusium. (e,f). Cells of pseudostipe. (g) Volva hyphae. (h) Rhizomorph hyphae. (i) Basidiospores. Scale bars: a = 20 mm, b, g = 10 μm, c, h, i = 5 μm, d = 20 μm, e–f = 50 μm.
Basionym: Dictyophora merulina Berk. (1886)
Synonyms:
≡ Clautriavia merulina (Berk.) Lloyd (1909)
= Dictyophora irpicina Pat. (1898)
= Phallus irpicinus (Pat.) Lloyd (1907)
Notes on morphology from Thai specimens: Egg globose to subglobose, 40–50 mm in diam., dark grayish yellowish brown to light gray (RHS2015 N200A to N200D) with a white mycelial rhizomorph arising from the base. Exoperidium papery, light brownish gray (RHS2015 N200C); mesoperidium gelatinous or lightly viscous, transparent to subtransparent, 3–5 mm thick, dark grayish yellow (RHS2015 N199D); endoperidium membranous, thin, white (RHS2015 N155D), covering the upper surface of gleba. Mature basidiomata 120–160 mm high. Cap campanulate, incurved toward the pseudostipe, surface very densely and merulioid-wrinkled, sticky, 10–30 mm high, 10–30 mm wide, light yellow or moderate yellow (RHS2015 160B or 161A), apex round to truncate with an apical pore. Gleba light olive brown (RHS2015 199B), mucilaginous. Pseudostipe 100–160 mm high, cylindrical, tapering, 13–35 mm wide at the base, 10–25 mm wide at the apex, white (RHS2015 NN155D), fragile, soft and spongy, hollow. Indusium coarsely latticed, white (RHS2015 NN155D), extended to ⅓ the size of the pseudostipe. The meshes of the indusium are large, polyhedral to round, 2–5 mm wide, the upper meshes larger than the lower meshes, the lower mesh margin wavy and thin. Volva subglobose, incurved toward the pseudostipe, 40–50 mm in diam., light brownish gray (RHS2015 200C), scar surface, with mycelial rhizomorphs from the base. Odor fetid.
Basidiospores 3.5–4.5 × 1.2–1.5 µm (x̅ = 4.2 × 1.3 μm, n = 25), subcylindrical to long–ellipsoid, subhyaline, smooth, thin-walled. Cap cells and hyphae; cells 10–50 µm in diam., globose to subglobose, hyaline, thin-walled; hyphae 2–3 μm wide, hyaline, thin-walled, septate, branched with clamp connections. Cell of pseudostipe 22–63 µm in diam., globose to subglobose, hyaline, smooth surface, thin-walled, bubble-like. Cell of indusium 20–57 µm in diam., globose to subglobose, hyaline, smooth surface, thin-walled, bubble-like. Volva hyphae; outer layer 3.75–10.0 μm wide, pale brown to brown, branched, smooth surface, thin-walled, septate with clamp connections; inner layer 1.2–7.5(20) μm wide, hyaline, branched, smooth surface, thin-walled, septate with clamp connections, swollen at the tip. Rhizomorph hyphae; outer layer 2.5–7.5 µm wide, hyaline to a pale brown, septate, branched, smooth surface, thick-walled with clamp connections; inner layer 1.2–7.5 µm wide, hyaline, branched, septate, smooth surface, thin-walled with clamp connections.
Known distribution: Australia [Citation31], Brazil [Citation32], China [Citation33], French Guiana [Citation34], India [Citation35,Citation36], Indonesia [Citation37–42], Philippines [Citation43], Republic of Trinidad and Tobago [Citation18], Sri Lanka [Citation44–48], Thailand [Citation18].
Habit and Habitat: on decomposing rice straw.
Culture characteristics: Tissue germinated on PDA within 24 h. Colonies were grown on PDA, immersed mycelium, reaching 2 cm in diam. in 1 month at 25 °C, surface and reverse white to cream.
Materials examined: THAILAND, Samut Sakhon Province, on decomposing rice straw, September 7 2019, U. Pinruan, (BBH 47826, BCC 92056; BBH 49055, BCC 92057; BBH 49057; BBH 49058, BBH 49059, BBH 49060, BBH 49061, BBH 49062, BBH 49063, BBH 49064, BBH 49065, BBH 49066).
Notes: The combined sequences of Phallus merulinus (BCC 92056 and BCC 92057) are identical to those of P. merulinus (INPA 240010), with 100% BSMP, 100% BSML, and 1.00 BPP. Phallus merulinus is the most distinctive among the Phallus species, mainly due to its cap, which has merulioid-wrinkled on the surface, and pale volva whereas most Phallus species have conspicuously reticulate indusium and dark volva.
Discussion
At present, most taxonomic studies of Phallus have been based on morphological features and molecular analyses. In this study, we introduce a new species, Phallus chiangmaiensis, based on its unique macro- and micro- morphological characteristics together with the support of molecular phylogenetic analyses. This species is morphologically related to the well-known species, P. indusiatus (Vent.) Desv. They have a campanulate cap with a reticulated surface and a pore at the apex. Their gleba are mucilaginous and moderately olive-brown. They have a white pseudostipe and a well-developed net-like, white indusium without a serrated margin. Their volva are non-echinulate and have rhizomorphs. However, spores of the new species are greenish-white, while those of P. indusiatus are hyaline. Phallus indusiatus has smaller basidiocarps and capsizes than those of the new species (basidiocarps: 15–20 mm; cap sizes: 18–32 × 16–27 mm). The pseudostipe and indusium length of P. indusiatus are shorter than those of the new species (pseudostipe: 75–110 × 11–22 mm; indusium: 100–200 mm) [Citation17]. Moreover, the new species has caps covered with a greenish white membrane and hyaline basidia, which are not observed in P. indusiatus. Apart from the morphology, the molecular phylogenetic analysis revealed that the two species were separate.
In the molecular phylogenetic analyses, which were based on sequences of the ITS, LSU, and atp6 gene regions, this species was well separated from other Phallus species with high bootstrap support values (). The most closely related species in the phylogenetic trees are P. echinovolvatus, P. fuscoechinovolvatus, P. multicolor (Berk. & Broome) Cooke, and P. luteus. Phallus echinovolvatus and P. fuscoechinovolvatus are similar to the new species in having a long white indusium, but they differ in having echinulate volva [Citation11,Citation17,Citation49,Citation50]. Phallus chiangmaiensis always has non-echinulate and milk white volva, that is never in blackish or black color. Phallus multicolor has a lemon yellow to yellowish orange indusium and a yellowish white pseudostipe [Citation51,Citation52] while the new species has white indusium and pseudostipe which differs. Phallus luteus has a yellowish orange indusium and a pale pink to reddish purple volva [Citation14] while the new species differs in having white indusium and milk white volva. Their morphological comparison is summarized in .
The findings of our study appear to be represented a re-encounter of Phallus merulinus 93 years after its first record in Thailand. Both the macroscopic and microscopic features of our specimens agree well with previous descriptions [Citation18,Citation34,Citation36]. The molecular phylogenetic trees revealed that four sequences of P. merulinus were related to P. atrovolvatus. Morphologically, P. merulinus is similar to P. atrovolvatus in having a rugulose to merulioid cap surface and white indusium. They have closely sizes of spores. However, the volva of P. atrovolvatus appears to be globose to ovoid shapes (19–47 × 18–29 mm) and always blackish color [Citation53,Citation54], while volva shapes of P. merulinus are globose to subglobose, and slightly larger (40–50 mm in diam.), and the volva color always paler [Citation34,Citation52,Citation55]. Phallus merulinus can be also separated from P. atrovolvatus by the unpleasant smell of the gleba (odor fetid) while the gleba odor of the latter species are strong, sweet, and aromatic (but never fetid) [Citation54].
Several species of Phallus are frequently reported in Thailand (some species reported as Dictyophora) [Citation55–60]. However, P. merulinus was not included in those reports which are considered rare species. The first report in Thailand of the species is in 1977 by Reid [Citation18], and this is the second report.
Dictyophora or Phallus were morphologically separated by the presence or absence of indusium. However, recent molecular phylogenetic analyses revealed most Dictyophora species belong to Phallus [Citation16,Citation49,Citation61–63]. In this study, the phylogenetic analyses () also show that Phallus species with indusium (P. atrovolvatus, P. chiangmaiensis, P. cinnabarinus, P. denigricans, P. echinovolvatus, P. fuscoechinovolvatus, P. haitangensis, P. indusiatus, P. lutescens, P. luteus, P. merulinus, P. multicolor, P. purpurascens, P. rubrovolvatus, P. serratus, P. squamulosus, P. ultraduplicatus) and without (P. calongei, P. campanulatus, P. coronatus, P. dongsun, P. flavocostatus, P. hadriani, P. impudicus, P. mengsongensis, P. ravenelii, P. rugulosus) are mixed together. Whether Dictyophora is a valid genus or conspecific with Phallus can only be verified if the type of the genus (D. phalloidea from Guiana) has been sequenced and compared with Phallus. The only sequence of D. phalloidea from GenBank is an ITS sequence of a specimen from Korea which grouped with P. rubrovolvatus [Citation17].
Supplemental Material
Download MS Word (539.6 KB)Acknowledgements
We would like to thank the project “Shaping modern fungal taxonomy – an integrative approach using morpho-molecular characterization, chemotaxonomy, and Next Generation Sequencing of invertebrate-pathogenic fungi and edible mushrooms” from BIOTEC, Grants P-19-50231. We also thank Dr. Theerayut Toojinda for his continued interest and support. Thanks, Mr. Siriwit Saliprasertchok and Ms. Chutchanun Trakulnaleamsai for helping us to collect the specimens.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Arora D. Mushrooms demystified: a comprehensive guide to the fleshy fungi. Berkeley: Ten Speed Press; 1986. p. 1056.
- Kreisel H. A preliminary survey of the genus Phallus sensu lato. Czech Mycol. 1996;48(4):273–281.
- Liu B, Fan L, Li JZ, et al. Flora fungorum sinicorum. Vol. 23. Beijing: Science Press; 2005. p. 137–171.
- Dai YC, Yang ZL. A revised checklist of medicinal fungi in China. Mycosystema. 2008;27:801–824.
- Dai YC, Zhou LW, Yang ZL, et al. A revised checklist of edible fungi in China. Mycosystema. 2010;29:1–21.
- Wu F, Zhou LW, Yang ZL, et al. Resource diversity of Chinese macrofungi: edible, medicinal and poisonous species. Fungal Divers. 2019;98(1):1–76.
- Li T, Li TH, Deng W, et al. Phallus dongsun and P. lutescens, two new species of Phallaceae (Basidiomycota) from China. Phytotaxa. 2020;443(1):19–037.
- Chaiyama V, Mau JL, Keawsompong S. Morphological characteristics, molecular identification and antioxidant activities of Phallus atrovolvatus (Agaricomycetes) isolated from Thailand. Int J Med Mushrooms. 2020;22(8):743–753.
- Dai YC, Yang ZL, Cui BK, et al. Species diversity and utilization of medicinal mushrooms and fungi in China (review). Int J Med Mushr. 2009;11(3):287–302.
- Calonge FD, Kreisel H. Phallus minusculus sp. nova from tropical Africa. Feddes Repert. 2002;113(7–8):600–602.
- Kreisel H, Hausknecht A. The gasteral Basidiomycetes of Mascarenes and Seychelles 3. Some recent records. Österr Z Pilzk. 2002;18:149–159.
- Baseia IG, Gibertoni TB, Maia LC. Phallus pygmaeus, a new minute species from a Brazilian tropical rain Forest. Mycotaxon. 2003;85:77–79.
- Calonge FD, de Sequeira M, Freitas T, et al. Phallus maderensis sp. nov., found in Madeira. Portugal. Bol Soc Micol Madrid. 2008;32:101–104.
- Kasuya T. Phallus luteus comb. nov. a new taxonomic treatment of a tropical phalloid fungus. Mycotaxon. 2008;106:7–13.
- Desjardin DE, Perry BA. A new species of Phallus from São Tomé, Africa. Mycologia. 2009;101(4):545–547.
- Li HL, Mortimer PE, Karunarathna SC, et al. New species of Phallus from a subtropical forest in xishuangbanna, China. Phytotaxa. 2014;163(2):91–103.
- Li HL, Ma XL, Mortimer PE, et al. Phallus haitangensis, a new species of stinkhorn from Yunnan province, China. Phytotaxa. 2016;280(2):116–128.
- Reid DA. Some Gasteromycetes from Trinidad and Tobago. Kew Bull. 1977;31(3):657–690.
- Hosaka K. Preliminary list of Phallales (Phallomycetidae, Basidiomycota) in Thailand. Mem Natl Mus Nat Sci. 2012;48:81–89.
- The Royal Horticultural Society. RHS colour chart, sixth revised edition. London; 2015.
- O'Donnell K, Cigelnik E, Weber NS, et al. Phylogenetic relationship among ascomycetous truffle and the true and false morels inferred from 18S and 28S ribosomal DNA sequence analysis. Mycologia. 1997;89(1):48–65.
- White TF, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA., Gelfand DH, Sninsky FS, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego (CA): Academic Press; 1990. p. 315–322.
- Sakayaroj J. Phylogenetics relationships of marine Ascomycota Ph.D. Thesis, Prince of Songkla University, Thailand; 2005.
- Raspé O, Vadthanarat S, Kesel A, et al. Pulveroboletus fragrans, a new Boletaceae species from Northern Thailand, with a remarkable aromatic odor. Mycol Prog. 2016;15:38.
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98.
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797.
- Miller M, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop 2010 (GCE), New Orleans, Louisiana, November 2010. p. 1–8.
- Swofford DL. PAUP: phylogenetic analysis using parsimony, version 4.0b10. Sunderland (MA): Sinauer Associates, Inc. Publishers; 2002.
- Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755.
- Cabral TS, Silva BD, Martín MP, et al. Behind the veil–exploring the diversity in Phallus indusiatus s.l. (Phallomycetidae, Basidiomycota). MycoKeys. 2019;58:103–127.
- Cooke MC. Australian fungi. Grevillea. 1882;11(58):57–65.
- Cabral TS, Clement CR, Baseia IG. Amazonian phalloids: new records for Brazil and South america. Mycotaxon. 2015;130(2):315–320.
- Liu B. The Gasteromycetes of China. Beih. Nova Hedwigia. 1984;76:1–235.
- Cheype JL. Phallaceae et clathrus récoltés en guyane française. Bull Mycol Bot Dauphiné-Savoie. 2010;197:51–66.
- Narasimhan MJ. The Phalloideae of Mysore. J Indian Bot Soc. 1932;11:248–254.
- Sridhar KR, Karun NC. On the basket stinkhorn mushroom Phallus merulinus (Phallaceae) in Mangalore, Karnataka, India. J Threat Taxa. 2013;5(5):3985–3988.
- Berkeley MJ. Egg fungi. Intellectual Observer. 1866;9:404.
- Patouillard N. Quelques champignons de java. Bull Trimest Soc Mycol Fr. 1898;14:182–198.
- Penzig O. Ueber javanische phalloideen. Ann Jard Bot Buitenzorg. 1899;16:133–173.
- Fischer E. Untersuchungen zur vergleichenden entwicklungsgeschichte und systematik der phalloideen. Denkschriften Der Schweizerischen Naturforschenden Gesellschaft. 1900;36:1–84.
- Lloyd CG. The phalloids of Australasia. Mycol Writings. 1907;2:1–24.
- Boedijn KB. The Phallineae of The Netherlands East Indies. Bull Jard Bot Buitenzorg Series III. 1932;12:71–103.
- Kobayasi Y. On the genus Dictyophora, especially in the East-Asiatic group. Trans Br Mycol Soc Japan. 1965;6:1–10.
- Petch T. The Phalloideae of ceylon. Ann Roy Bot Gard. 1908;4:139–184.
- Lloyd CG. Mycological notes no. 32. Mycol Writings. 1909;3:413–424.
- Lloyd CG. Synopsis of the known Phalloids. Mycological Notes No. 3. Mycol Writings. 1909;3:1–96.
- Lloyd CG. Mycological notes no. 34. Mycol Writings. 1910;3:449–450.
- Lloyd CG. Mycological notes no. 35. Mycol Writings. 1910;3:461–476.
- Zang M, Zheng D-R, Hu Z-X. A new species of the genus Dictyophora from China. Mycotaxon. 1988;33:145–148.
- Song B, Li T, Li TH, et al. Phallus fuscoechinovolvatus (Phallaceae, Basidiomycota), a new species with a dark spinose volva from Southern China. Phytotaxa. 2018;334(1):19–27.
- Berkeley MJ, Broome CE. List of fungi from Brisbane, Queensland; with descriptions of new species. Part II. Trans Linn Soc Lond. 1883;2:53–73.
- Cunningham GH. The Gasteromycetes of Australia and New Zealand. Dunedin: McIndoe; 1994. p. 236.
- Calonge FD, Kreisel H, Mata M. Phallus atrovolvatus, a new species from Costa Rica. Bol Soc Micol Madrid. 2005;29:5–8.
- Das K, Hembrom ME, Parihar A. Two interesting species of stinkhorns from India. NeBIO. 2013;4(4):1–6.
- Soytong K. Mushrooms and macrofungi in Thailand. Ubonratchathani: Siritham Offset Publishers Ltd; 1994. p. 222.
- Ruksawong P, Flegel TW. Thai mushrooms and other fungi. Bangkok: National Center for Genetic Engineering and Biotechnology and National Science and Technology Development Agency, BIOTEC; 2001. p. 268.
- Chandrasrikul A, Suwanarit P, Sangwanit U, et al. Diversity of mushrooms and macrofungi in Thailand. Bangkok: Kasetsart University Press; 2008. p. 514.
- Sanoamuang N. Wild mushrooms of Thailand: biodiversity and utilization. Bangkok: Universal Graphic and Trading Co., Ltd; 2010. p. 424.
- Chandrasrikul A, Suwanarit P, Sangwanit U, et al. Checklist of mushrooms (Basidiomycetes) in Thailand. Bangkok: Office of Natural Resources and Environmental Policy and Planning; 2013. p. 448.
- Sangwanit U, Suwanarit P, Payappanon A, et al. Checklists of biological properties: Mushrooms. Bangkok: Biodiversity-Based Economy Development Office (Public Organization); 2013. p. 374.
- Calonge FD. A tentative key to identify the species of Phallus. Bol Soc Micol Madrid. 2005;29:9–17.
- Baseia IG, Maia LC, Calonge FD. Notes on Phalalles in the neotropics. Bol Soc Micol Madrid. 2006;30:87–93.
- Moreno G, Khalid AN, Alvarado P, et al. Phallus hadriani and P. roseus from Pakistan. Mycotaxon. 2013;125(1):45–51.
