Abstract
The truffle and ectomycorrhizal roots formed by Tuber sp. were collected from the rhizosphere of Quercus aliena in Korea. The morphological characteristics of the ascoma, and molecular phylogenetic analysis using sequences from the internal transcribed spacer (ITS) and large subunit (LSU) of ribosomal DNA, translation elongation factor 1-alpha (TEF), and RNA polymerase second largest subunit (RPB2) regions confirmed the distinct morphology of the truffle. This truffle belongs to a monophyletic clade among the other Tuber species in the phylogeny. This study describes the truffle, Tuber koreanum, as a new species reported from Korea.
1. Introduction
The genus Tuber P. Micheli ex F. H. Wigg. belongs to the family Tuberaceae, order Pezizales, and division Ascomycota. Tuber species produce hypogenous fruiting bodies called truffles. Truffles have several ascospores in a hyaline ascus, and the ascospores have an alveolate–reticulate or a spino-reticulate shaped surface [Citation1]. Some truffles are edible and have commercial value because of their unique aroma [Citation2]. Tuber species have an ectomycorrhizal (ECM) relationship with the roots of some woody plants belonging to the families Betulaceae, Cistaceae, Corylaceae, Fagaceae, and Pinaceae [Citation3,Citation4].
More than 180 Tuber species have been recorded in the MycoBank database (http://mycobank.org) and new species are continuously being reported worldwide [Citation5]. However, Tuber species have been poorly studied and only five species have been reported till date in Korea: T. aestivum subsp. uncinatum, T. borchii, T. himalayense, T. huidongense, and T. indicum [Citation6–9]. During a survey of Tuber species in Korea, Tuber ascoma were collected from the rhizosphere of oak trees. Based on their morphological characteristics and molecular analysis, the ascoma did not represent any previously described species of the genus Tuber. Therefore, in this study, we report a novel Tuber species from Korea.
2. Materials and methods
2.1. Sampling
Fresh specimens of fruiting bodies and ECM roots were collected from the rhizosphere of oak trees, Quercus aliena Blume., in Gyeongju (N35°43'10.8", E129°20'36.3") in September 2020. The specimens were transported to the laboratory, where the morphological characteristics of ascoma and ascospores were observed under a dissecting microscope and an optical microscope. The ascocarps examined were deposited in the Herbarium of Korea National University of Education, Cheongju, Korea.
2.2. Morphological analysis
Mycorrhizal roots were observed under a microscope, and the mycorrhizal cross sections were obtained using a microtome (Leica Microsystems Nussloch GmbH, Nussloch, Germany). Morphological characteristics such as the color, branching systems, texture, and hyphae of the mycorrhiza were recorded [Citation10].
2.3. Phylogenetic analysis
The genomic DNA of the ECM root tips and ascomata were extracted and amplified using PCR. ITS1F/ITS4 primers were used to amplify the internal transcribed spacer (ITS) region of ribosomal DNA (rDNA) [Citation11]. In addition, LR0R/LR16 primers to amplify the large subunit (LSU) region[Citation12], EF1α Tuber-f/EF1α Tuber-r that amplifies translation elongation factor 1-alpha (TEF) region [Citation5], and fRPB2-5F/fRPB2-7cR primers that amplify RNA polymerase second largest subunit (RPB2) region [Citation13] were used for ascomata. After PCR, each DNA band was confirmed through polymerase gel electrophoresis and sent for DNA sequencing (SolGent Co. Ltd., Daejeon, Korea). The DNA sequence information was used to find similarities using the BLAST search program from the National Center for Biological Information (NCBI). The phylogenetic trees of aligned DNA sequences () were constructed using maximum likelihood (ML) method by Kimura 2-parameter (K2P) substitution model [Citation14]. The ambiguous characters were excluded from the analysis, and the gaps were treated as missing data. The ML bootstrap replicates (1000) were computed for the best scoring ML tree. Bayesian analysis [Citation15] was performed for calculating posterior probabilities (>50% majority rule consensus trees) on the branches of phylogenetic tree.
Table 1. List of Tuber sequences used in the phylogenetic analysis.
3. Results and discussion
3.1. Taxonomy
Tuber koreanum H. Park & A. H. Eom, sp. nov. [#MB840072] (.
Figure 1. Morphological characteristics of Tuber koreanum. Ascoma (A, B), gleba and peridium (C), asci (D), and ascospores (E, F) (scale bars: B, C = 1000 μm, D = 100 μm, E, F = 10 μm).
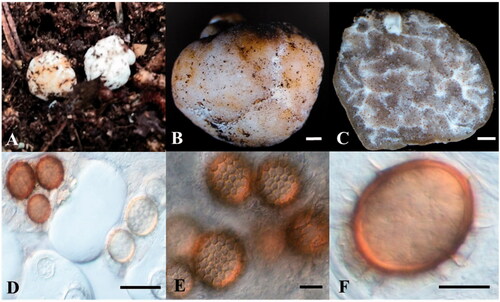
Type: KOREA. Gyeongsangbukdo: Gyeongju-si, in rhizosphere soil under Quercus aliena, 9 September 2020, collected by Hyeok Park, GB20004 (holotype), GB20011 and GB20046 (isotype). GenBank no.: OK275104 (ITS), OK275105 (LSU).
Etymology: The first new truffle species discovered in Korea
ASCOMATA globose to subglobose, rarely ovoid, irregularly rugged surface, bright white to yellowish beige, (5–20) × (4–15) mm in diameter. GLEBA grayish brown to yellowish brown, bright white mycelia mixed in partially, pale brown in the part with mature ascospores. PERIDIUM (61.0–)71.2(–92.8) μm in thickness, dark beige to grayish brown, divided into two layers, outer layer yellowish brown and thicker than inner layer, inner layer mucoid, dark brown, white mycelia distributed in several places. ASCI hyaline, squashed, ellipsoid to conical, with smooth margin, aseptate, size differing based on the number of ascospores (2–4 spores) in each ascus, (22.6–)38.5(–51.2) × (20.1–)29.8(–34.8) μm in diameter. ASCOSPORE initially bright ivory or pale gold, reddish brown to pale brown in mature spore, glittering, subglobose to ovoid, sometimes ellipsoid (13.2–)17.2(–21.9) × (13.2–)15.9(–19.6) μm in diameter. The polygonal pieces on the surface of the mature ascospore form a reticulate ornament with several spines on the external surface; spines are usually curved, with sharp end, (1.9–)2.7(–3.4) μm high.
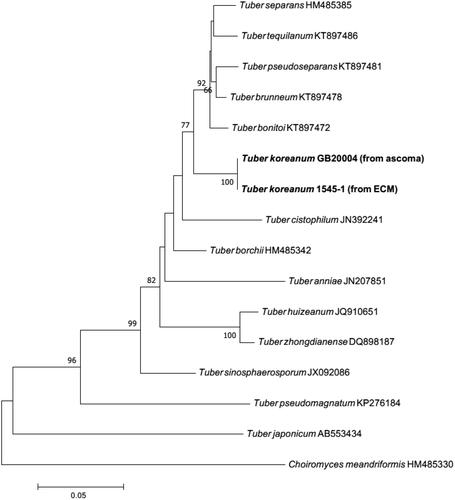
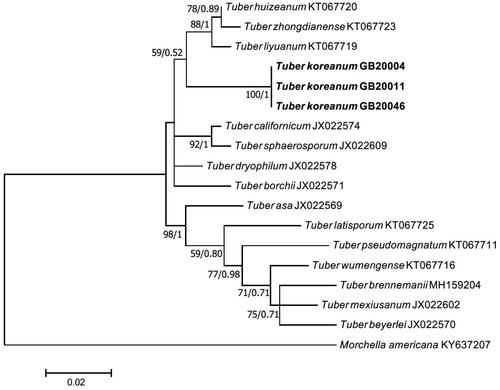
Mycorrhiza: The ECOM root tips were straight, rarely curved, pale yellow to ivory, vertically branched, and the length of the branch from the side was generally shorter than that of the forward branch (). The fungal mantle layer showed an interlocking-synenchyma structure (). The ITS rDNA sequence of ECM root tip showed a coincidence with the sequences of the ascoma and the mycelium. Furthermore, they formed a monophyletic group on the phylogenetic tree ().
Figure 2. Morphological characteristics of ectomycorrhiza colonized by Tuber koreanum from root of Quercus aliena. Mycorrhizal root tips (A, B); Fungal mantle layer (E) (scale bars: A, B = 500 μm, C, D = 20 μm).
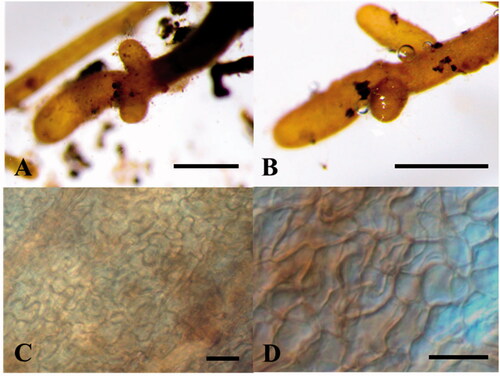
Figure 3. Maximum-likelihood phylogenetic tree of Tuber koreanum based on the alignment of the internal transcribed spacer (ITS) rDNA sequences obtained from ectomycorrhizal root tip. Choiromyces meandriformis was used as an outgroup. Numbers on branches indicate bootstrap values (1,000 replicates). Sequences from the present study were in bold.
The morphological characteristics of GB20004 ascoma were compared with the other truffle species that have similar appearances (). The GB20004 ascoma have beige or light white peridium, light grayish-brown or yellowish-brown gleba, and conical or irregular oval ascus, whereas Tuber borchii showed reddish-brown or dark brown peridium, dark brown gleba, and an oval-shaped ascus tapering at the base [Citation16]. Some characteristics of GB20004 ascoma were similar to Tuber flavidosporum or Tuber japonicum, a truffle discovered in Japan [Citation17]. The ascospores of GB20004 were mainly ovoid, and sometimes ellipsoid, while the ascospores of T. flavidosporum or T. japonicum were close to perfect globose shaped. The color of GB20004 ascospores was ivory or light gold, initially but changed to reddish-brown or pale brown as the spores matured. T. flavidosporum and T. japonicum showed white or pale yellow ascospores [Citation17]. In addition, the peridium thickness of T. flavidosporum and T. japonicum was more than 200 μm [Citation17], while that of GB20004 was less than 100 μm. GB20004 showed morphological characteristics that were clearly distinguishable from T. borchii, T. flavidosporum, and T. japonicum.
Table 2. Morphological characteristics of Tuber koreanum with the allied Tuber species.
On the ML phylogenetic tree, GB20004 formed a monophyletic group distinct from other species. The analysis of the ITS-LSU region and the TEF DNA sequences showed that GB20004 was closer to Tuber liyuanum, Tuber huizeanum, and Tuber zhongdianense than to T. borchii, T. flavidosporum and T. japonicum ( and ). An analysis of the RPB2 DNA sequence also showed that GB20004 was clearly located apart from T. borchii, T. flavidosporum and T. japonicum (), implying that GB20004 is distinctly different from these three species both morphologically and molecularly. Based on these results, we determined Tuber koreanum GB20004 as a novel truffle species that has not been recorded yet.
Figure 4. Phylogenetic tree of Tuber koreanum ascoma inferred using the maximum likelihood method based on alignment of ITS and LSU DNA sequences. Venturia pyrina was used as an outgroup. Strains used in this study are in bold.
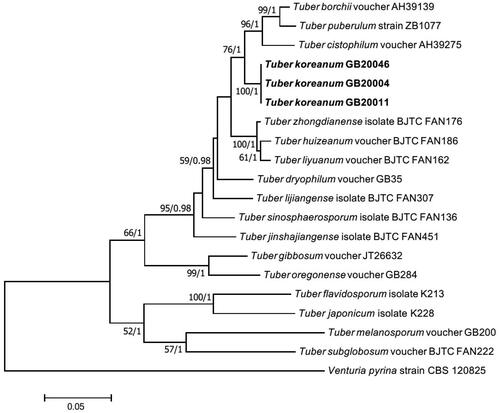
Figure 5. Phylogenetic tree of Tuber koreanum ascoma inferred using the maximum likelihood method based on alignment of TEF DNA sequences. Bootstrap values and Bayesian posterior probabilities are indicated below branches. Epicoccum latusicollum was used as an outgroup. Strains used in this study are in bold.
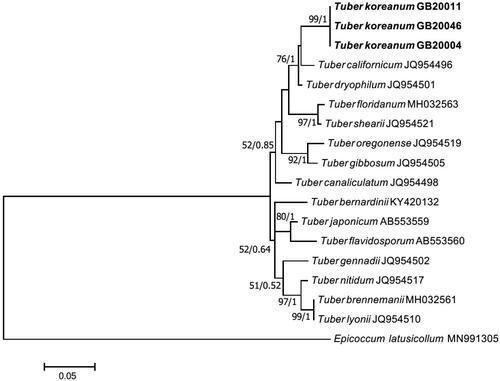
Figure 6. Phylogenetic tree of Tuber koreanum ascoma inferred using the maximum likelihood method based on alignment of RPB2 DNA sequences. Bootstrap values and Bayesian posterior probabilities are indicated below branches. Morchella americana was used as an outgroup. Strains used in this study are in bold.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bonito G, Gryganskyi A, Trappe J, et al. A global meta-analysis of Tuber ITS rDNA sequences: species diversity, host associations and long-distance dispersal. Mol Ecol. 2010;19(22):4994–5008.
- Trappe JM. The orders, families, and genera of hypogeous Ascomycotina (truffles and their relatives). Mycotaxon. 1979;9:297–340.
- Bulman SR, Visnovsky SB, Hall IR, et al. Molecular and morphological identification of truffle-producing Tuber species in New Zealand. Mycol Prog. 2010;9(2):205–214.
- Pacioni G, Comandini O. Tuber. Ectomycorrhizal fungi key genera in profile. In: Cairney WG, Chambers SM, editors. Berlin, Germany: Springer Verlag; 1999. p. 163.
- Bonito G, Smith ME, Nowak M, et al. Historical biogeography and diversification of truffles in the Tuberaceae and their newly identified Southern hemisphere sister lineage. PLoS One. 2013;8(1):e52765.
- Shin KS, Park JS, Yoshimi S. Note on Tuber aestivum subsp. uncinatum newly recorded in Korea. Kor J Mycol. 1995;23:10–13.
- Park H, Gwon JH, Lee JC, et al. Report on Tuber huidongense, a truffle species previously unrecorded in Korea. Kor J Mycol. 2020;48:505–510.
- Park H, Gwon JH, Lee JC, et al. Morphological and phylogenetic characteristics of Tuber himalayense collected from rhizosphere of Quercus dentata in Korea. Kor J Mycol. 2021;49:101–108.
- Seok SJ, Lim YW, Kim CM, et al. List of mushrooms in Korea. Seoul: The Korean Society of Mycology; 2013.
- Goodman D, Durall D, Trofymow J, et al. Concise descriptions of some North American ectomycorrhizae. Victoria, BC: Victoria: Canada-BC Forest Resource Development Agreement, Canadian Forest Service; 1996–1999.
- Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2(2):113–118.
- Moncalvo JM, Lutzoni FM, Rehner SA, et al. Phylogenetic relationships of agaric fungi based on nuclear large subunit ribosomal DNA sequences. Syst Biol. 2000;49(2):278–305.
- Liu YJ, Whelen S, Hall BD. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol Biol Evol. 1999;16(12):1799–1808.
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874.
- Drummond AJ, Suchard MA, Xie D, et al. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29(8):1969–1973.
- Elliot TF, Türkoğlu A, Trappe JM, et al. Turkish truffles 2: eight new records from Anatolia. Mycotaxon. 2016;131(2):439–453.
- Kinoshita A, Sasaki H, Nara K. Two new truffle species, Tuber japonicum and Tuber flavidosporum spp. nov. found from Japan. Mycoscience. 2016;57(5):366–373.
