Abstract
During a survey of fungal diversity associated with insects, mud, soil, and freshwater niches in different areas in Korea, nine interesting fungal strains were isolated. Based on their morphological characteristics and molecular phylogeny analyses, using a combined data set of β-tubulin (BenA), calmodulin (CaM), and second largest subunit of RNA polymerase (RPB2) sequences, the strains CNUFC AM-44, CNUFC JCW3-4, CNUFC S708, CNUFC WT202, CNUFC AS1-29, CNUFC JCW3-5, CNUFC JDP37, and CNUFC JDP62 were identified as Aspergillus alabamensis, A. floridensis, A. subversicolor, Penicillium flavigenum, P. laevigatum, P. lenticrescens, Talaromyces adpressus, and T. beijingensis, respectively. The strain CNUFC JT1301 belongs to series Westlingiorum in section Citrina and is phylogenetically related to P. manginii. However, slow growth when cultivated on CYA, MEA, CREA is observed and the property can be used to easily distinguish the new species from these species. Additionally, P. manginii is known to produce sclerotia, while CNUFC JT1301 strain does not. Herein, the new fungal species is proposed as P. aquadulcis sp. nov. Eight species, A. alabamensis, A. floridensis, A. subversicolor, P. flavigenum, P. laevigatum, P. lenticrescens, T. adpressus, and T. beijingensis, have not been previously reported in Korea. The present study expands the known distribution of fungal species belonging to the families Aspergillaceae and Trichocomaceae in Korea.
1. Introduction
Aspergillus, Penicillium, and Talaromyces are well-known species of the Aspergillaceae and Trichocomaceae families, have a worldwide distribution, and have been isolated from a diverse range of habitats from air to soil, to vegetation and indoor environments [Citation1–3]. Many species of Aspergillus, Penicillium, and Talaromyces are important both in industry and medicine, whereas others are associated with food spoilage, mycotoxin production, and dangerous human and plant diseases [Citation4,Citation5]. Various species can grow at low or high temperatures, low pH values, high salt or sugar concentrations, or low oxygen levels [Citation6,Citation7].
The genus Aspergillus was first introduced by Micheli in 1729 [Citation8] to describe asexual fungi whose conidiophores resembled an aspergillum. Later, in 1768, Haller [Citation9] validated the genus; in 1832, Fries [Citation10] sanctioned the generic name, with Aspergillus glaucus (L.) Link [= Mucor glaucus L. ≡ Monilia glauca (L.) Pers.] as the generic type. Members of this genus produce many different compounds, which are used or have potential to be used in pharmacology and biotechnology [Citation1,Citation5]. Aspergillus is subdivided into 6 subgenera, 27 sections, and 75 series [Citation11]. Currently, this genus consists of 446 species [Citation11].
In addition, in 1809, Link [Citation12] established the genus Penicillium, with the generic type P. expansum, to accommodate asexual fungi that bore Penicillium (painter’s brush)-like fruiting bodies. They are also known for their ability to produce antibiotics, organic acids, alcohols, enzymes, pharmaceuticals, and other metabolites including mycotoxins [Citation2,Citation4,Citation6]. This genus is divided into 2 subgenera, 32 sections, and 89 series [Citation11]. Currently, the genus contains 483 accepted species [Citation11].
The genus Talaromyces was established by Benjamin (1955) [Citation13] for a teleomorph of Penicillium, with Talaromyces vermiculatus (=T. flavus) as the type species. These species are characterized by cleistothecial or gymnothecial ascomata, unitunicate 8-spored asci, and unicellular ascospores with or without equatorial crests. The anamorphs have predominantly biverticillate or rarely terverticillate conidiophores with acerose phialides and narrow column [Citation3,Citation14]. The genus is divided into seven sections based on multigene phylogenetic, morphological, and physiological characteristics reported by Yilmaz et al. [Citation3]. Recently, a new section Tenues was proposed by Sun et al. [Citation15]. Currently, the genus contains 171 accepted species [Citation11]. Although the number of new species in the genera Aspergillus, Penicillium, and Talaromyces has increased in Korea, this number is significantly lower than the global number of accepted species. In addition, some habitats such as freshwater and insect hosts are still poorly studied.
The objective of this study was to morphologically and molecularly characterize of a new species, P. aquadulcis, and eight previously unrecorded fungal species in Korea—namely A. alabamensis, A. floridensis, A. subversicolor, P. flavigenum, P. laevigatum, P. lenticrescens, T. adpressus, and T. beijingensis—isolated from freshwater, insects, mud, and soil.
2. Materials and methods
2.1. Sample collection and isolation
Freshwater, insect, mud and soil samples were collected from different areas in Korea. The samples were collected in sterile plastic bags or sterile 50 mL Falcon tubes and transferred to the laboratory. To isolate fungi from freshwater, soil, and mud, a serial dilution plating method was used. Briefly, 1 mL of water or 1 g of soil or mud was mixed with 9 mL of sterile distilled water; serial dilutions ranging from 10−1 to 10−5 were prepared. An aliquot of 0.1 mL from each dilution was transferred to potato dextrose agar (PDA; Difco, Becton, Dickinson and Co., Sparks, MD, USA) or yeast malt extract agar (YMA; Difco) supplemented with the antibiotic streptomycin sulfate (100 ppm) and was incubated at 25 °C for 3–7 days. To isolate pure cultures, individual colonies with various morphologies were picked, transferred to PDA, and subcultured until pure mycelia were obtained. Fungal isolation from insect samples were conducted following our previous methods [Citation16]. All pure isolates were maintained in PDA slant tubes and in 20% glycerol at −80 °C at the Environmental Microbiology Laboratory Fungarium, Chonnam National University, Gwangju, Korea as CNUFC AM-44, CNUFC JCW3-4, CNUFC S708, CNUFC JT1301, CNUFC WT202, CNUFC AS1-29, CNUFC JCW3-5, CNUFC JDP37, and CNUFC JDP62. The isolates were also deposited at the Collection of National Institute of Biological Resources (NIBR) Incheon, and Nakdonggang National Institute of Biological Resources (NNIBR), Sangju, Korea. Type specimens were deposited in Chonnam National University Herbarium, Gwangju, Korea as inactive dried cultures. Information on all isolates used in this study is shown in .
Table 1. Source information and GenBank accession numbers of strains used in this study.
2.2. Morphological characteristics
The strains were three-point inoculated onto Czapek yeast autolysate agar (CYA; yeast extract, Difco), malt extract agar (MEA; Difco), yeast extract sucrose agar (YES; yeast extract, Difco), except creatinine sucrose agar (CREA) only for CNUFC JT1301 strain. All petri dishes were inoculated at 25 °C for 7 days. Medium preparation, inoculation, and incubation were performed according to the methods reported by Yilmaz et al. [Citation3]. Colony characteristics were recorded after 7 days. Lactic acid (60%) was used as the mount fluid, and 96% ethanol was used to remove excess conidia. The cultures were examined and photographed under an Olympus BX53 light microscope with an attached Olympus DP74 camera (Olympus, Tokyo, Japan). For SEM, samples were performed as described previously by Nguyen et al. [Citation17]. The samples were then sputter-coated with gold and imaged with scanning electron microscope (Hitachi S4700 Field-Emission SEM, Hitachi, Tokyo, Japan) at the Korea Basic Science Institute, Gwangju, Korea.
2.3. DNA extraction, PCR, and sequencing
Fungal isolates were cultured on PDA overlaid with cellophane at 25 °C for 5–7 days. Genomic DNA was extracted using the Solg TM Genomic DNA Preparation Kit (Solgent Co. Ltd., Daejeon, Korea). The internal transcribed spacer (ITS) region was amplified using the primer pairs V9G and ITS4 [Citation18,Citation19]. β-tubulin (BenA) was amplified using the primer pairs T10 and Bt2b [Citation20]. Calmodulin (CaM) was amplified using the primer pairs CF1 and CF4 [Citation21,Citation22]. Second largest subunit of RNA polymerase (RPB2) was amplified using the primer pairs RPB2-5F and RPB2-7cR [Citation23]. PCR amplification was performed according to the conditions described by Visagie et al. [Citation2] and Yilmaz et al. [Citation3]. The PCR products were then purified using an Accuprep PCR Purification Kit (Bioneer Corp., Daejeon, Korea) and sequenced on the ABI PRISM 3730XL Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) using the same primers employed for PCR.
2.4. Phylogenetic analysis
Sequences for the selected strains were retrieved from the GenBank and aligned using MAFFT (http://mafft.cbrc.jp/alignment/server) with the algorithm L-INS-I. Aligned sequences were automatically trimmed using trimAl [Citation24] using the gappyout method. Concatenated datasets of the BenA, CaM and RPB2 genetic markers were built using MEGA7 [Citation25]. Phylogenetic reconstructions by maximum likelihood (ML) was carried out using RAxML-HPC2 on XSEDE on the online CIPRES Portal (https://www.phylo.org/portal2) with bootstrap support obtained from 1,000 replicates. The GTR + GAMMA model of nucleotide evolution was used. Bayesian inference analysis was performed with MrBayes 3.2.2 [Citation26], using a Markov Chain Monte Carlo (MCMC) algorithm. The sample frequency was set to 100 and the first 25% of trees were removed as burn-in. Support values are provided at the branches [ML bootstrap support (MLBS), BI posterior probability (BPP)]. Hamigera avellanea CBS 295.48 was chosen as the outgroup in the Aspergillus and Penicillium phylogenies. Penicillium corylophilum CBS 312.48 was chosen as the outgroup in the section Citrina. Talaromyces varians CBS 386.48 was the outgroup for the phylogeny of Talaromyces. The newly obtained sequences have been deposited in the GenBank database under the accession numbers provided in .
3. Results
3.1. Molecular phylogeny
Combined BenA, CaM and RPB2 was used to clarify the phylogenetic relationships of newly collected specimens with other species in the genera Aspergillus, Penicillium, and Talaromyces ().
Figure 1. Phylogram generated from the Maximum Likelihood (RAxML) analysis based on a combined BenA, CaM and RPB2 sequence data for species classified in the Aspergillus sections Nigri, Terrei and Nidulantes. Bootstrap values ≥70% MLBS and ≥0.90 BPP are indicated above or below branches. Hamigera avellanea CBS 295.48 was used as the outgroup. The newly generated sequences are indicated in blue bold. T = ex type.
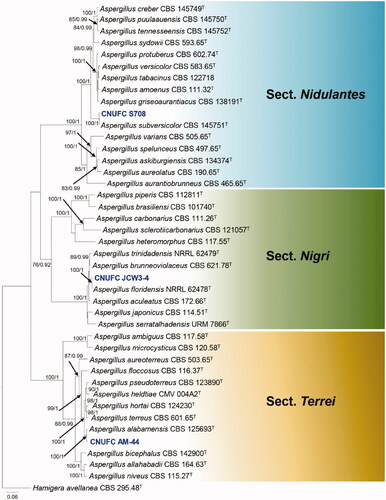
Figure 2. Phylogram generated from the Maximum Likelihood (RAxML) analysis based on a combined BenA, CaM and RPB2 sequence data for species classified in Penicillium section Citrina. Bootstrap values ≥70% MLBS and ≥0.90 BPP are indicated above or below branches. Penicillium corylophilum CBS 312.48 was used as outgroup. The newly generated sequence is indicated in blue bold. T = ex type.
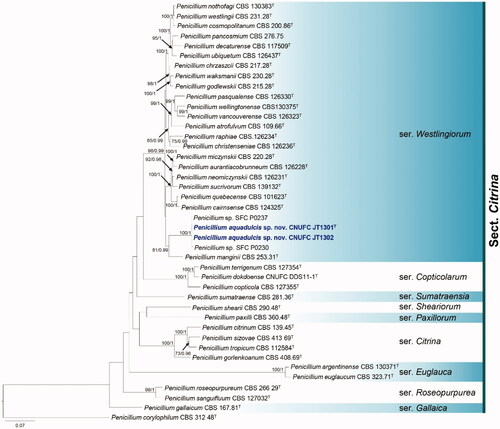
Figure 3. Phylogram generated from the Maximum Likelihood (RAxML) analysis based on a combined BenA, CaM and RPB2 sequence data for species classified in the Penicillium sections Chrysogena, Lanata-Divaricata and Ramosum. Bootstrap values ≥70% MLBS and ≥0.90 BPP are indicated above or below branches. Hamigera avellanea CBS 295.48 was used as the outgroup. The newly generated sequences are indicated in blue bold. T = ex type.
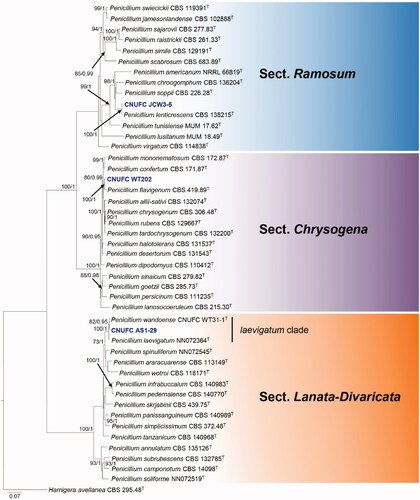
Figure 4. Phylogram generated from the Maximum Likelihood (RAxML) analysis based on a combined BenA, CaM and RPB2 sequence data for species classified in the section Talaromyces. Bootstrap values ≥70% MLBS and ≥0.90 BPP are indicated above or below branches. Talaromyces varians CBS 386.48 was used as the outgroup. The newly generated sequences are indicated in blue bold. T = ex type.
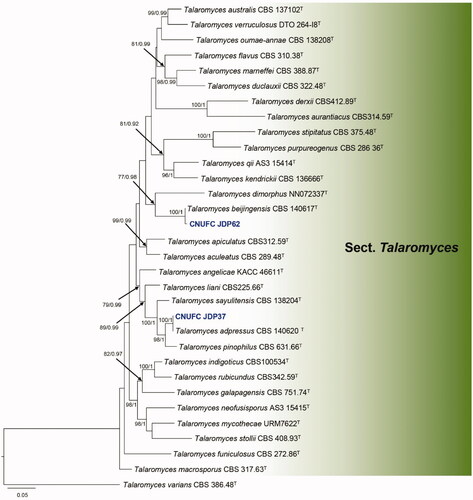
The three-locus phylogenetic tree with the 41 isolates of Aspergillus is shown in . The concatenated alignment consists of 2,227 characters (including alignment gaps): 541, 676, and 1,010 characters used in the BenA, CaM, and RPB2, respectively. The isolates CNUFC S708, CNUFC JCW3-4, and CNUFC AM-44 are clustered within the same clade as Aspergillus subversicolor CBS 145751 (ex-type strain), A. floridensis NRRL 62478 (ex-type strain), and A. alabamensis CBS 125693 (ex-type strain), in the sections Nidulantes, Nigri, and Terrei, respectively.
The phylogenetic trees generated by the concatenated BenA-CaM-RPB2 sequences (424, 528, 915 and 1,867 characters with gaps, respectively) indicates that our two isolates CNUFC JT1301 and CNUFC JT1302 are related to P. manginii with high statistical support ().
The phylogenetic tree based on the three-locus with the 45 isolates of Penicillium is shown in . The concatenated alignment consists of 1,918 characters (including alignment gaps): 422, 551, and 945 characters used in the BenA, CaM, and RPB2, respectively. The isolates CNUFC WT202, CNUFC AS1-29, CNUFC JCW3-5 are clustered within the same clade as P. flavigenum CBS 419.89, P. laevigatum NN072364, and P. lenticrescens CBS 138215, in the sections Chrysogena, Lanata-Divaricata, and Ramosum, respectively.
The phylogenetic tree based on the three-locus () shows the relationships of the two Talaromyces isolates included in the study. The concatenated alignment consists of 1,675 characters (including alignment gaps): 372, 465, and 838 characters used in the BenA, CaM, and RPB2, respectively. The isolates CNUFC JDP37 and CNUFC JDP62 are clustered within the same clade as T. adpressus CBS 140620 (ex-type strain) and T. beijingensis CBS 140617 (ex-type strain) in the section Talaromyces.
3.2. Taxonomy
3.2.1. Taxonomy of CNUFC AM-44
Aspergillus alabamensis Balajee, Baddley, Frisvad & Samson, Eukaryotic Cell 8(5): 720 (2009) ().
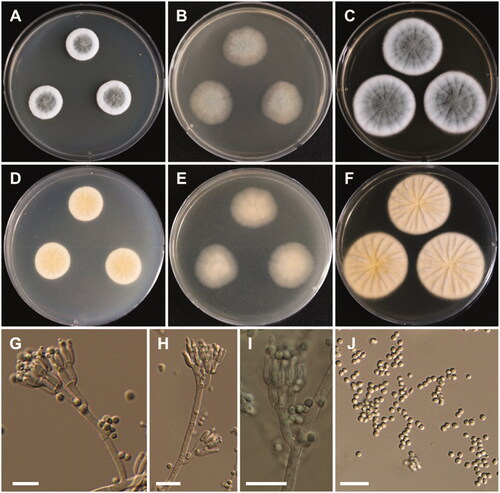
Figure 5. Morphology of Aspergillus alabamensis. A, D: Colonies on Czapek yeast autolysate agar (CYA). B, E: Colonies on malt extract agar (MEA). C, F: Colonies on Yeast extract sucrose agar (YES). (A–C: obverse view, D–F: reverse view). (G–I) Conidiophores; (J) Conidia (scale bars: G–I = 20 μm, J = 10 μm).
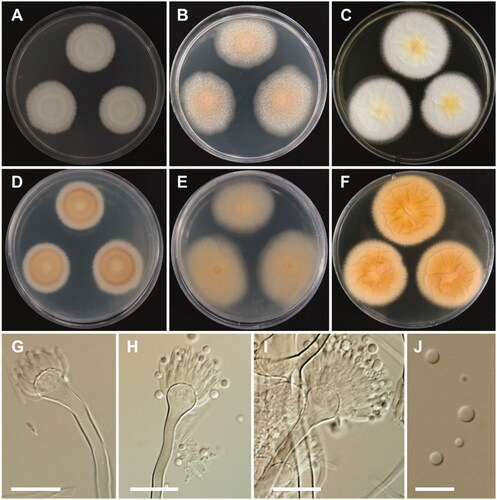
Colony characteristics: On CYA, colonies floccose, plane; mycelium white; sporulation moderate; soluble pigments absent; reverse pale yellowish brown; and reached 28–30 mm in diameter after 7 days at 25 °C. On MEA, colonies floccose, plane; mycelium white to yellowish brown at the center; sporulation moderate to dense; soluble pigments absent; reverse pale-yellow brown; and reached 33–35 mm in diameter after 7 days at 25 °C. On YES, colonies plane, slightly sulcate; mycelium white; sporulation in the colony center; soluble pigments absent; reverse pale orange; and reached 31–33 mm in diameter after 7 days at 25 °C.
Micromorphology: Conidiophores biseriate, smooth-walled, stipes 3.5–7 µm. Vesicles subglobose, 6–24 µm in diameter. Metulae closely packed, 5.5–10.5 × 2–3 µm. Phialides ampulliform, 5.5–9.5 × 1.5–3 µm. Conidia globose to subglobose, smooth-walled, 2–3 µm.
Strain examined: REPUBLIC OF KOREA, Anmyeon-eup (36°34'46.0"N, 126°19'42.7"E), Taean-gun, Chungnam Province, from mud sample, 29 January 2019, H.B. Lee (culture CNUFC AM-44).
Note: CNUFC AM-44 is morphologically similar to Aspergillus alabamensis described by Balajee et al. [Citation27]. However, CNUFC AM-44 showed to have a partly consistent vesicle size (6–24 µm) that is larger than that of A. alabamensis CBS 125639 (10–16 µm) [Citation27]. Moreover, colony color of CNUFC AM-44 observed on CYA was white, whereas that of CBS 125639 (ex-type strain) was yellowish- to cinnamon-brown [Citation27]. Aspergillus alabamensis was isolated from clinical specimens from Alabama, USA [Citation27], and soil samples from Cuba, Argentina, New Mexico, USA, and China [Citation28]. The strain isolated in this study was found in mud.
3.2.2. Taxonomy of CNUFC JCW3-4
Aspergillus floridensis Jurjević, G. Perrone & S.W. Peterson, IMA Fungus 3(2): 169 (2012) ().
Figure 6. Morphology of Aspergillus floridensis. A, D: Colonies on Czapek yeast autolysate agar (CYA). B, E: Colonies on malt extract agar (MEA). C, F: Colonies on yeast extract sucrose agar (YES). (A–C: obverse view, D–F: reverse view). (G–I) Conidiophores. (J) Conidia (scale bars: G–I = 20 μm, J = 10 μm).
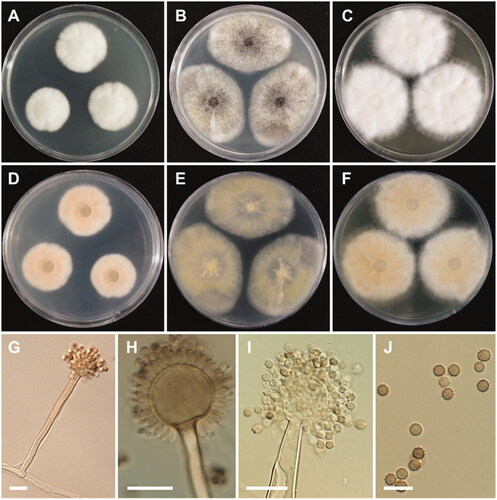
Colony characteristics: On CYA, colonies floccose, moderately radially sulcate; mycelium white; sporulation poor; soluble pigments absent; reverse pale orange; and reached 24–29 mm in diameter after 7 days at 25 °C. On MEA, colonies floccose, plane; mycelium white to grayish-yellow; conidial heads abundant; sporulation absent; soluble pigments absent; reverse pale yellow; and reached 36–44 mm in diameter after 7 days at 25 °C. On YES, colonies floccose, radially sulcate toward the center; mycelium white; sporulation poor; soluble pigments absent; reverse dull white to pale yellow, and reached 33–41 mm in diameter after 7 days at 25 °C.
Micromorphology: Conidiophores uniseriate, smooth-walled, stipes 7.5–15 µm. Vesicles globose to subglobose, (8.5–)23.5–50 µm in diameter. Phialides cover entire vesicle, 5.5–8.5 × 3–4.5 µm. Conidia globose to ellipsoidal, with finely spiny to echinulate surface, and brown at maturity, 3–4.5 µm.
Strain examined: REPUBLIC OF KOREA, Cheonjeyeon (33°15'02.5"N, 126°24'59.6"E) located in Jeju Island, Jeju Province, from waterfall freshwater sample, 18 April 2019, H.B. Lee (culture: CNUFC JCW3-4).
Note: CNUFC JCW3-4 also has similar morphological characteristics with the previously described species of A. floridensis [Citation29]. However, the vesicles and conidia are slightly smaller than those of. A. floridensis [Citation29] [Vesicles: (14−)35–65(−105) µm diam; conidia: 4–5(−6)×3.5–5.5 µm]. In addition, the colony diameter of CNUFC JCW3-4 (24–31 mm) was shorter than that of A. floridensis NRRL 62478 (80–85 mm) on CYA [Citation29].
3.2.3. Taxonomy of CNUFC S708
Aspergillus subversicolor Jurjević, S.W. Peterson & B.W. Horn, IMA Fungus 3(1): 72 (2012). ().
Figure 7. Morphology of Aspergillus subversicolor. A, D: Colonies on Czapek yeast autolysate agar (CYA). B, E: Colonies on malt extract agar (MEA). C, F: Colonies on yeast extract sucrose agar (YES). (A–C: obverse view, D–F: reverse view). (G–I) Conidiophores; (J) Conidia (scale bars: G–J = 20 μm).
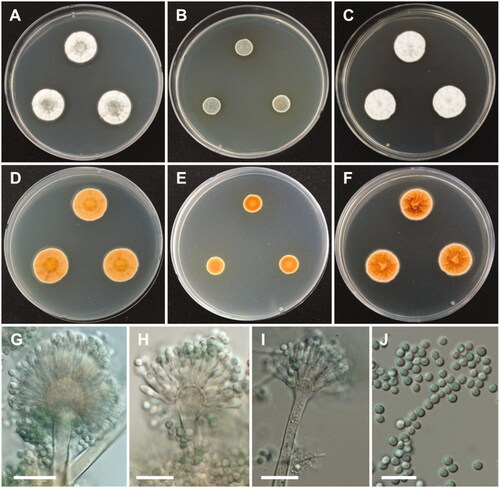
Colony characteristics: On CYA, colonies plane; mycelium white to gray green at the center, wrinkled, radially sulcate toward the center; sporulation sparse; soluble pigments absent; reverse honey yellow; and reached 20–22 mm in diameter after 7 days at 25 °C. On MEA, colonies plane; mycelium green; sporulation bluish green; soluble pigments absent; reverse brownish orange, and reached 10–12 mm in diameter after 7 days at 25 °C. On YES, colonies plane, radially sulcate; mycelium pale white; sporulation absent; soluble pigments absent; reverse deep orange; and reached 20–23 mm in diameter after 7 days at 25 °C.
Micromorphology: Conidiophores biseriate, smooth-walled, stipes 4–6.5 µm. Vesicles pyriform or subglobose, 8–17.5(–21.5) µm in diameter. Metulae, 5–8 × 2.5–4. Phialides ampulliform, 5.5–8 (–9)×2–3.5 µm. Conidia globose to subglobose, green, 2.5–3.5 µm.
Strain examined: REPUBLIC OF KOREA, Kunryang-ri (36°44’00.2″N, 126°78’18.0″E), Cheongyang-eup, Cheongyang, Chungnam Province, from the head of bees, 7 May 2020, H.B. Lee (culture: CNUFC S708).
Note: The morphological descriptions of CNUFC S708 were matched well with the previously described species of A. subversicolor [Citation30]. However, our observations showed that the size of conidia of isolate CNUFC S708 were smaller than previously reported [Citation30]. Colony diameter on CYA media was shown to be different from the previously described A. subversicolor (CYA: 22–25 mm after 10 days at 25 °C).
3.2.4. Taxonomy of CNUFC JT1301
Penicillium aquadulcis Hyang B. Lee & T.T.T. Nguyen, sp. nov.
Index Fungorum: IF556953; .
Figure 8. Morphology of Penicillium aquadulcis sp. nov. A, E: Colonies on Czapek yeast autolysate agar (CYA). B, F: Colonies on yeast extract sucrose agar (YES). C, G: Colonies on malt extract agar (MEA). D, H: Colonies on creatine sucrose agar (CREA). (A–D: obverse view, E–H: reverse view), (I–L: LM; M, N: SEM). (I–K, M) Conidiophores. (L, N) Conidia (scale bars: I–K, M = 10 μm, L, N = 5 μm).
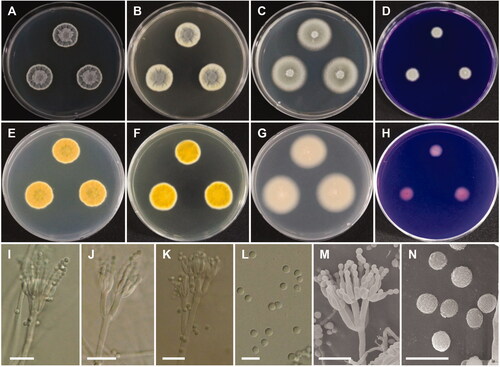
Etymology: aquadulcis. Referring to the freshwater source from which the species was first isolated.
Type specimen: REPUBLIC OF KOREA, Cheonjeyeon (33°15'02.5"N, 126°24'59.6"E) located in Jeju Island, Jeju Province, from waterfall freshwater sample, 18 April 2019, H.B. Lee (CNUFC HT19009 holotype; ex-type culture: CNUFC JT1301).
Colony diameter after 7 days at 25 °C, in mm: CYA: 16–20 mm; MEA: 27–30; YES: 16–22; CREA: 5–8. CYA, 15 °C: 21–24; CYA, 30 °C: 14–16; CYA, 37 °C: no growth.
Colony characters: CYA, 25 °C, 7 days: Colonies elevated in center; radial sulcate, deep; margin entire to slightly irregular; mycelium white; colony texture floccose in center; sporulation moderate; exudate absent; soluble pigment absent; reverse yellow amber. YES, 25 °C, 7 days: Colonies radially sulcate; margins entire; mycelium light yellow; colony texture floccose at center; sporulation moderate to good, mainly in center; exudate absent; soluble pigment absent; reverse yellow brown. MEA, 25 °C, 7 days: Colonies plane, slightly raised at center; margins entire; mycelium white; colony texture velvety, slightly floccose in center; sporulation strong; exudate absent; soluble pigment absent; reverse light brown. CREA, 25 °C, 7 days: weak growth, no acid production.
Micromorphology: Sclerotia absent. Conidiophores mostly biverticillate, sometimes terverticillate or monoverticillate, with round or smooth walled stipes, non-vesiculate, stipe 2.5–3.5 µm wide. Metulae 2–6 per stipe, 8.5–12.5(–16)×2.5–3.5 µm. Phialides flask-shaped, 3–10 per metula, 7–10.5 × 2–3 µm. Conidia in short chains, finely rough walled, globose to subglobose, 2–2.5 µm.
Note: Our phylogenetic analyses showed that P. aquadulcis forms a separate lineage, phylogenetically related to P. manginii. However, slow growth when cultivated on CYA, MEA, CREA is observed and the property can be used to easily distinguish the new species from these species. The conidial shape of P. aquadulcis (globose to subglobose) is different from that of P. manginii (ellipsoidal). Additionally, P. manginii is known to produce sclerotia, while P. aquadulcis does not.
Additional material examined: REPUBLIC OF KOREA, Cheonjeyeon (33°15'02.5"N, 126°24'59.6"E) located in Jeju Island, Jeju Province, from waterfall freshwater sample, 18 April 2019, H.B. Lee (culture: CNUFC JT1302).
3.2.5. Taxonomy of CNUFC WT202
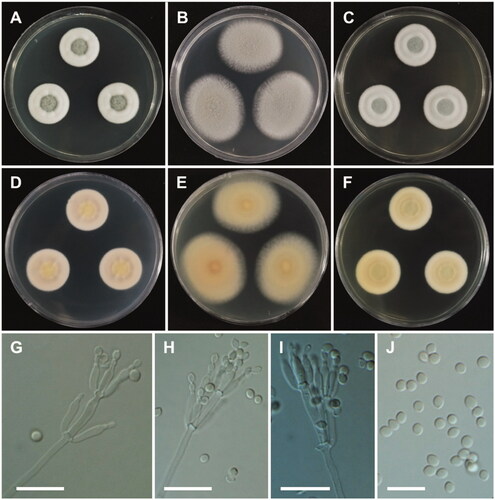
Penicillium flavigenum Frisvad &Samson, Mycological Research 101(5): 622 (1997) ().
Figure 9. Morphology of Penicillium flavigenum. A, D: Colonies on Czapek yeast autolysate agar (CYA). B, E: Colonies on malt extract agar (MEA). C, F: Colonies on yeast extract sucrose agar (YES). (A–C: obverse view, D–F: reverse view). (G–I) Conidiophores; (J) Conidia (scale bars: G–J = 20 μm).
Colony characteristics: On CYA, colonies floccose, radially sulcate; mycelium white; exudate absent; soluble pigments absent; sporulation moderate; reverse pale yellow; and reached 19–21 mm in diameter after 7 days at 25 °C. On MEA, colonies plane; exudate absent; soluble pigments absent; sporulation strong; reverse light yellow; and reached 24–27 mm in diameter after 7 days at 25 °C. On YES, colonies plane, radially sulcate; mycelium white; exudate absent; soluble pigments absent; sporulation moderate; reverse pale yellow; and reached 35–38 mm in diameter after 7 days at 25 °C.
Micromorphology: Conidiophores terverticillate, stipes, 2–5 µm. Metulae were cylindrical, 7–11 × 2.5–3.5 µm. Phialides were cylindrical, 5.5–8.5 × 2–3 µm. Conidia smooth-walled, globose to subglobose, 2.5–3.5 µm.
Strain examined: REPUBLIC OF KOREA, Kunryang-ri (36°44'00.2"N, 126°78'18.0"E), Cheongyang-eup, Cheongyang, Chungnam Province, from freshwater sample, 1 June 2020, H.B. Lee (culture: CNUFC WT202).
Note: There were differences observed in the colony diameter for CNUFC WT202 in comparison to previous descriptions. Its colony diameter was different from that of previously described Penicillium flavigenum on CYA (21–31 mm) [Citation31]. In this isolate, conidia were slightly smaller than Penicillium flavigenum (2.5–4 µm) [Citation31].
3.2.6. Taxonomy of CNUFC AS1-29
Penicillium laevigatum L. Cai, Houbraken, R.N. Barbosa & X.Z. Jiang, Cladistics 35(5): 537 (2018) ().
Figure 10. Morphology of Penicillium laevigatum. A, D: Colonies on Czapek yeast autolysate agar (CYA). B, E: Colonies on malt extract agar (MEA). C, F: Colonies on yeast extract sucrose agar (YES). (A–C: obverse view, D–F: reverse view). (G–I) Conidiophores; (J) Conidia (scale bars: G–I = 20 μm, J = 10 μm).
Colony characteristics: On CYA, colonies slightly sulcate; margins entire; mycelium white; sporulation sparse; soluble pigments absent; exudates absent; reverse light grayish yellow; and reached 21–23 mm in diameter after 7 days at 25 °C. On MEA, mycelium white; slightly raised at the center; sporulation strong; soluble pigments absent; exudates absent; reverse light orange yellow; and reached 36–40 mm in diameter after 7 days at 25 °C. On YES, colonies slightly raised at the center; margins entire; mycelium white; sporulation sparse; soluble pigments absent; exudates absent; reverse buff-yellow; and reached 23–25 mm in diameter after 7 days at 25 °C.
Micromorphology: Conidiophores mono and biverticillate, stipes, 2–4 µm. Metulae 2–3 per stipe, appressed to divergent, 8.5–19 × 2–4 µm. Phialides ampulliform, 6–17 × 2–3.5 µm. Conidia smooth-walled, globose to subglobose, 2–3 µm.
Strain examined: REPUBLIC OF KOREA, Anmyeon-eup (36°34'46.0"N, 126°19'42.7"E), Taean-gun, Chungnam Province, from soil sample, 29 January 2019, H.B. Lee (culture CNUFC AS1-29).
Note: Based on phylogenetic analysis of combined BenA, CaM, and RPB2 sequence data, our isolate CNUFC AS1-29 was clustered with the ex-type strains of P. laevigatum and P. wandoense in laevigatum clade (). The colony morphology and culture characteristics of CNUFC AS1-29 strain were similar to the previous descriptions [Citation32]. As reported by Houbraken et al. [Citation11], P. laevigatum and P. wandoense, which were isolated from freshwater collected at Wando, Korea [Citation33], were conspecific. However, P. wandoense (3–5 µm) differs from P. laevigatum (1–14 µm) by having smaller number of phialides per metulae [Citation32]. Penicillium wandoense also differs from P. laevigatum (type species) and isolate CNUFC AS1-29 in terms of reverse color on MEA. Thus, future morphology and molecular studies are needed to confirm the relationship between P. laevigatum and P. wandoense.
3.2.7. Taxonomy of CNUFC JCW3-5
Penicillium lenticrescens Visagie, Seifert & Samson, Studies in Mycology 78: 123 (2014) ().
Figure 11. Morphology of Penicillium lenticrescens. A, D: Colonies on Czapek yeast autolysate agar (CYA). B, E: Colonies on malt extract agar (MEA). C, F: Colonies on yeast extract sucrose agar (YES). (A–C: obverse view, D–F: reverse view). (G, H) Conidiophores; (I) Conidia (scale bars: G, H = 20, I = 10 μm).
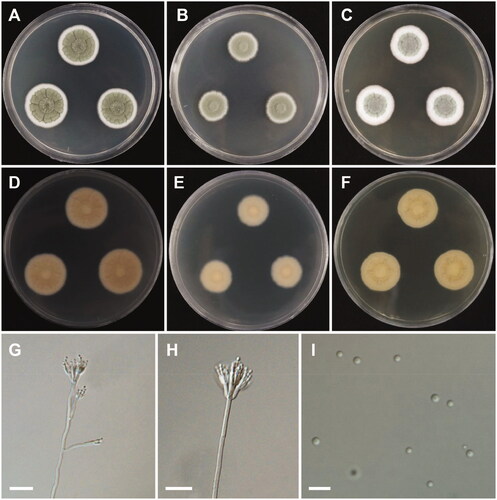
Colony characteristics: On CYA, colonies sulcate; margins entire; mycelia white; sporulation moderate; soluble pigments absent; exudates absent; reverse pale yellow; and reached 23–29 mm in diameter after 7 days at 25 °C. On MEA, colonies slightly raised at the center, plane; mycelium white; soluble pigments absent; exudates absent; reverse yellowish gray; and reached 16–18 mm in diameter after 7 days at 25 °C. On YES, colonies sulcate; margins entire; mycelia white; slightly raised at the center; sporulation absent; soluble pigments absent; exudates absent; reverse greenish yellow; and reached 19–23 mm in diameter after 7 days at 25 °C.
Micromorphology: Conidiophores biverticillate and divaricate, stipes smooth, 2.5–3.5 µm. Metulae divergent, 8.5–14.5 × 2–4 µm. Phialides ampulliform, 7.5–12 × 2–3.5 µm. Conidia globose to subglobose, 2–3 µm.
Strain examined: REPUBLIC OF KOREA, Cheonjeyeon (33°15'02.5"N, 126°24'59.6"E) located in Jeju Island, Jeju Province, from waterfall freshwater sample, 18 April 2019, H.B. Lee (culture: CNUFC JCW3-5).
Note: The morphological characteristics of the isolate were consistent with those of P. lenticrescens, as described by Visagie et al. [Citation2]. However, our strain produced biverticillate and divaricate conidiophores, in contrast to the mostly biverticillate sporangiophores of P. lenticrescens described by Visagie et al. [Citation2].
3.2.8. Taxonomy of CNUFC JDP37
Talaromyces adpressus A.J. Chen, Frisvad & Samson, Studies in Mycology 84: 124 (2016) ().
Figure 12. Morphology of Talaromyces adpressus. A, D: Colonies on Czapek yeast autolysate agar (CYA). B, E: Colonies on malt extract agar (MEA). C, F: Colonies on yeast extract sucrose agar (YES). (A–C: obverse view, D–F: reverse view). (G–I) Conidiophores; (J) Conidia (scale bars: G–J = 10 μm).
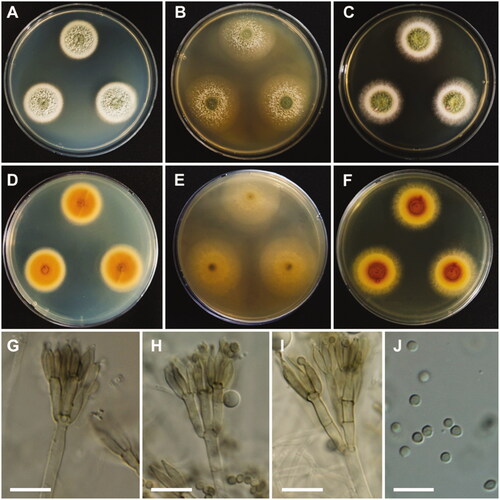
Colony characteristics: On CYA, colonies slightly sulcate; margins entire; mycelium white; texture floccose; sporulation moderately to dense; soluble pigments absent; exudates absent; reverse yellowish orange, and reached 22–24 mm in diameter after 7 days at 25 °C. On MEA, colonies slightly raised at the center, plane; mycelium white; sporulation sparse; texture floccose in the center; soluble pigments absent; exudates absent; reverse light yellow; and reached 32–34 mm in diameter after 7 days at 25 °C. On YES, colonies raised at the center; appearance green; margins entire; mycelium white; sporulation moderate; soluble pigments absent; exudates absent; reverse brownish orange and light yellow; and reached 28–30 mm in diameter after 7 days at 25 °C.
Micromorphology: Conidiophores biverticillate, stipes smooth, 2.5–4 µm. Metulae 2–5, 9–16 × 2.5–3.5 µm. Phialides 2–5, 8.5–12 × 2–3 µm. Conidia smooth, globose to subglobose or ellipsoidal, 2–4 µm.
Strain examined: REPUBLIC OF KOREA, Mokpo (34°49'17.5"N, 126°24'35.0"E), Jeonnam province, from freshwater sample, September 2020, H.B. Lee (culture: CNUFC JDP37).
Note: The morphological characteristics of the isolate CNUFC JDP37 in this study were similar to those of the previously described T. adpressus by Chen et al. [Citation34]. However, our observations showed that the size of conidia of the isolate CNUFC JDP37 was smaller than that of previously reported species.
3.2.9. Taxonomy of CNUFC JDP62
Talaromyces beijingensis A.J. Chen, Frisvad & Samson, Studies in Mycology 84: 125 (2016) ().
Figure 13. Morphology of Talaromyces beijingensis. A, D: Colonies on Czapek yeast autolysate agar (CYA). B, E: Colonies on malt extract agar (MEA). C, F: Colonies on yeast extract sucrose agar (YES). (A–C: obverse view, D–F: reverse view). (G–I) Conidiophores; (J) Conidia (scale bars: G–J = 10 μm).
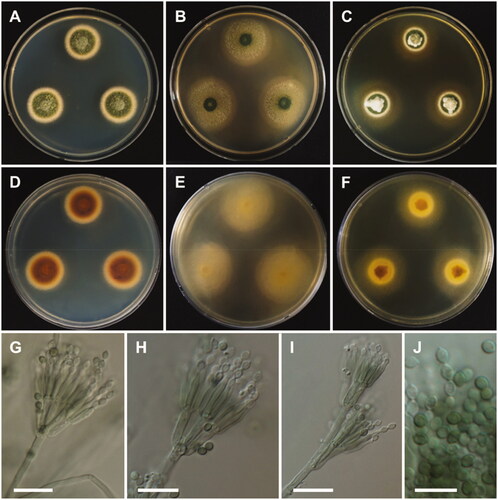
Colony characteristics: On CYA, colonies moderately deep, raised at the center; margins entire; mycelium white; texture velvety; sporulation dense; soluble pigments absent; reverse peach; and reached 26–29 mm in diameter after 7 days at 25 °C. On MEA, colonies slightly raised at the center, plane; mycelium white; sporulation sparse; reverse light yellow; and reached 31–33 mm in diameter after 7 days at 25 °C. On YES, colonies raised at the center; appearance green; margins entire; mycelium white; sporulation moderate; soluble pigments absent; exudates absent; reverse brownish orange at the center, yellow at the edge; and reached 29–31 mm in diameter after 7 days at 25 °C.
Micromorphology: Conidiophores mostly terverticillate, sometimes biverticillate, stipes smooth, 2.5–3.5 µm. Metulae 2–5, 10–14 × 2.5–3.5 µm. Phialides 9.5–13 × 2.0–3.0 µm. Conidia smooth, ellipsoidal to fusiform, 2.5–3.5 × 2–2.5 µm. Ascomata not observed.
Strain examined: REPUBLIC OF KOREA, Mokpo (34°49'17.5"N, 126°24'35.0"E), Jeonnam province, from freshwater sample, September 2020, H.B. Lee (culture: CNUFC JDP62).
Note: The morphological characteristics of the isolate CNUFC JDP62 in this study were similar to those of the previously described T. beijingensis by Chen et al. [Citation34]. However, the number of metulae are larger than those of T. beijingensis and smaller conidia [Citation34]. In addition, differences in colony diameter were observed on MEA (35–39 mm) [Citation34].
4. Discussion
Recent studies on the genera Aspergillus, Penicillium, and Talaromyces have been conducted in Korea, yet their distribution is not fully known, with various areas remain unexplored. In this study, a new species and eight new records are described and illustrated. The ITS is the DNA barcoding marker for the identification of fungi. However, many of the previous studies have shown that the only ITS is insufficient for Aspergillus, Penicillium, and Talaromyces identification as closely related species often share similar or identical sequences. The BenA, and CaM sequences have been used for the identification of these three genera, as this is the recommended identification marker [Citation2,Citation3,Citation11]. In this study, the multigene phylogeny proposed by Houbraken et al. [Citation11] allowed us to recognize a new species, and to identify eight recently described in the genera Aspergillus, Penicillium and Talaromyces. A new species described here belongs to section Citrina, series Westlingiorum. Penicillium aquadulcis is closely related to P. manginii, and are in one clade with Penicillium sp. SFC P0237 and Penicillium sp. SFC P0230. Penicillium sp. SFC P0237 and Penicillium sp. SFC P0230 were isolated from Rhododendron brachycarpum in Korea but there are no descriptions for these species [Citation35]. Penicillium aquadulcis differs from P. manginii by having smaller colonies on CYA, MEA, and CREA. Conidial shape and production of sclerotia can be easily used to distinguish between Penicillium aquadulcis and P. manginii. Section Citrina currently includes 41 species and 9 series (ser. Gallaica, ser. Roseopurpurea, ser. Euglauca, ser. Citrina, ser. Paxillorum, ser. Sheariorum, ser. Sumatraensia, ser. Copticolarum and ser. Westlingiorum) [Citation11]. Twenty-two species are currently accepted in series Westlingiorum and most of those species isolated from soil [Citation36]. In this study, we isolated a novel species from freshwater.
Hawksworth [Citation37] estimated that there are approximately 1.5 million fungal species on Earth. Around 3,000 species are known to be associated with aquatic habitats, with more than 600 species of ascomycetes reported from freshwater habitats [Citation38,Citation39]. The importance of these organisms in secondary metabolite production has also been recognized. However, the knowledge on freshwater fungi remains insufficient. Moreover, these species are less explored than terrestrial fungi.
In this study, both T. adpressus, and T. beijingensis were also found in freshwater. Talaromyces adpressus and T. beijingensis are known Talaromyces species, which were originally described from air in China. To the best of our knowledge, species from aquatic environments have been reported for the sections, Helici, Purpurei and Talaromyces [Citation40,Citation41]. Further studies are necessary to understand both the distribution and relevance of species in the section Talaromyces in aquatic environments.
Insects are known to host various fungi [Citation16,Citation42,Citation43]. Recently, Barbosa et al. [Citation42] reported an association of Penicillium and Talaromyces with stingless bees. Notably, we also isolated A. subversicolor from bee niche. Previous studies isolated A. subversicolor from coffee berries and as endophytes from macroalgae [Citation30,Citation44], suggesting that this species has a broad ecological niche. The occurrence and distribution of Aspergillus associated with insects in freshwater habitats remain unknown. Further studies on the relationship of fungi and insects are thus needed to understand the benefits that fungi may provide to their hosts. In addition, the genera Aspergillus, Penicillium, and Talaromyces are now known to produce secondary metabolites, warranting further studies on the biodiversity of these fungi and their potential applications in biotechnology.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Samson RA, Visagie CM, Houbraken J, et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud Mycol. 2014;78:141–173.
- Visagie CM, Hirooka Y, Tanney JB, et al. Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world. Stud Mycol. 2014;78:63–139.
- Yilmaz N, Visagie CM, Houbraken J, et al. Polyphasic taxonomy of the genus Talaromyces. Stud Mycol. 2014;78:175–341.
- Frisvad JC, Smedsgaard J, Larsen TO, et al. Mycotoxins, drugs and other extrolites produced by species in Penicillium subgenus Penicillium. Stud Mycol. 2004;49:201–241.
- Frisvad JC. Taxonomy, chemodiversity, and chemoconsistency of Aspergillus, Penicillium, and Talaromyces species. Front Microbiol. 2014;5:773.
- Houbraken J, Samson R. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stud Mycol. 2011;70(1):1–51.
- Houbraken J, Visagie CM, Meijer M, et al. A taxonomic and phylogenetic revision of Penicillium section Aspergilloides. Stud Mycol. 2014;78:373–451.
- Micheli PA. Nova plantarvm genera ivxta tovrnefortii methodvm disposita quibus plantae MDCCC recensentur. Florence:Typis B. Paperinii;1729.
- Haller A. von Historia stirpium indigenarum Helvetiae inchoate. Bernae. 1768.
- Fries EM. Systema mycologicum. E. Mauritius, Gryphiswald: Lundae; 1832.
- Houbraken J, Kocsubé S, Visagie CM, et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): an overview of families, genera, subgenera, sections, series and species. Stud Mycol. 2020;95:5–169.
- Link HF. Observationes in ordines plantarum naturales. Dissertatio 1ma. Magazin Der Gesellschaft Naturforschenden Freunde Berlin. 1809;3:3–42.
- Benjamin CR. Ascocarps of Aspergillus and Penicillium. Mycologia. 1955;47(5):669–687.
- Samson RA, Yilmaz N, Houbraken J, et al. Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Stud Mycol. 2011;70(1):159–184.
- Sun BD, Chen AJ, Houbraken J, et al. New section and species in Talaromyces. Mycokeys. 2020;68:75–113.
- Nguyen TTT, Voigt K, Santiago ALC, et al. Discovery of novel Backusella (Backusellaceae, Mucorales) isolated from invertebrates and toads in Cheongyang, Korea. JoF. 2021;7(7):513.
- Nguyen TTT, Paul NC, Lee HB. Characterization of Paecilomyces variotii and Talaromyces amestolkiae in Korea based on the morphological characteristics and multigene phylogenetic analyses. Mycobiology. 2016;44(4):248–259.
- de Hoog GS, Gerrits van den Ende AH. Gerrits van den ende AHG. Molecular diagnostics of clinical strains of filamentous basidiomycetes. Mycoses. 1998;41(5-6):183–189.
- White TJ, Bruns T, Lee S. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR protocols: a guide to methods and applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990. pp. 315–322.
- Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 1995;6:1323–1330.
- Peterson SW, Vega F, Posada F, et al. Penicillium coffeae, a new endophytic species isolated from a coffee plant and its phylogenetic relationship to P. fellutanum, P. thiersii and P. brocae based on parsimony analysis of multilocus DNA sequences. Mycologia. 2005;97(3):659–666.
- Hong SB, Cho HS, Shin HD, et al. Novel Neosartorya species isolated from soil in Korea. Int J Syst Evol Microbiol. 2006;56(2):477–486.
- Liu YJ, Whelen S, Hall BD. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol Biol Evol. 1999;16(12):1799–1808.
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25(15):1972–1973.
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874.
- Ronquist F, Teslenko M, van der Mark P, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542.
- Balajee SA, Baddley JW, Peterson SW, et al. Aspergillus alabamensis, a new clinically relevant species in the section Terrei. Eukaryot Cell. 2009;8(5):713–722.
- Samson RA, Peterson SW, Frisvad JC, et al. New species in Aspergillus section Terrei. Stud Mycol. 2011;69(1):39–55.
- Jurjević Z, Peterson SW, Stea G, et al. Two novel species of Aspergillus section Nigri from indoor air. IMA Fungus. 2012;3(2):159–173.
- Jurjević Z, Peterson SW, Horn BW. Aspergillus section Versicolores: nine new species and multilocus DNA sequence based phylogeny. IMA Fungus. 2012;3(1):59–79.
- Banke S, Frisvad JC, Rosendahl S. Taxonomy of Penicillium chrysogenum and related xerophilic species, based on isozyme analysis. Mycol Res. 1997;101(5):617–624.
- Diao YZ, Chen Q, Jiang XZ, et al. Penicillium section Lanata-Divaricata from acidic soil. Cladistics. 2019;35(5):514–549.
- Hyde KD, Tennakoon DS, Jeewon R, et al. Fungal diversity notes 1036–1150: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2019;96(1):1–242.
- Chen AJ, Sun BD, Houbraken J, et al. New Talaromyces species from indoor environments in China. Stud Mycol. 2016;84:119–144.
- Park MS, Lee JW, Kim SH, et al. Penicillium from rhizosphere soil in terrestrial and coastal environments in South Korea. Mycobiology. 2020;48(6):431–442.
- Houbraken J, Frisvad JC, Samson RA. Taxonomy of Penicillium section Citrina. Stud Mycol. 2011;70(1):53–138.
- Hawksworth DL. The fungal dimension of biodiversity: magnitude, significance, and conservation. Mycol Res. 1991;95(6):641–655.
- Shearer CA, Descals E, Kohlmeyer B, et al. Fungal biodiversity in aquatic habitats. Biodivers Conserv. 2007;16(1):49–67.
- Jones EBG, Hyde KD, Pang KL. Freshwater fungi and fungal-like organisms. Boston: De Gruyter; 2014.
- Peterson SW, Jurjević Ž. The Talaromyces pinophilus species complex. Fungal Biol. 2019;123(10):745–762.
- Nguyen TTT, Frisvad JC, Kirk PM, et al. Discovery and extrolite production of three new species of Talaromyces belonging to sections Helici and Purpurei from freshwater in Korea. JoF. 2021;7(9):722.
- Barbosa RN, Bezerra JDP, Souza-Motta CM, et al. New Penicillium and Talaromyces species from honey, pollen and nests of stingless bees. Antonie Van Leeuwenhoek. 2018;111(10):1883–1912.
- Nguyen TTT, Lee HB. Mucor cheongyangensis, a new species isolated from the surface of lycorma delicatula in Korea. Phytotaxa. 2020;446(1):33–42.
- Sarasan M, Job N, Puthumana J, et al. Exploration and profiling of hidden endophytic mycota of marine macroalgae with potential drug leads. FEMS Microbiol Lett. 2020;367:fnaa078.
