Abstract
The interaction of mating pheromone and pheromone receptor from the B mating-type locus is the first step in the activation of the mushroom mating signal transduction pathway. The B mating-type locus of Lentinula edodes is composed of Bα and Bβ subloci, each of which contains genes for mating pheromone and pheromone receptor. Allelic variations in both subloci generate multiple B mating-types through which L. edodes maintains genetic diversity. In addition to the B mating-type locus, our genomic sequence analysis revealed the presence of a novel chromosomal locus 43.3 kb away from the B mating-type locus, containing genes for a pair of mating pheromones (PHBN1 and PHBN2) and a pheromone receptor (RCBN). The new locus (Bα-N) was homologous to the Bα sublocus, but unlike the multiallelic Bα sublocus, it was highly conserved across the wild and cultivated strains. The interactions of RcbN with various mating pheromones from the B and Bα-N mating-type loci were investigated using yeast model that replaced endogenous yeast mating pheromone receptor STE2 with RCBN. The yeast mating signal transduction pathway was only activated in the presence of PHBN1 or PHBN2 in the RcbN producing yeast, indicating that RcbN interacts with self-pheromones (PHBN1 and PHBN2), not with pheromones from the B mating-type locus. The biological function of the Bα-N locus was suggested to control the expression of A mating-type genes, as evidenced by the increased expression of two A-genes HD1 and HD2 upon the treatment of synthetic PHBN1 and PHBN2 peptides to the monokaryotic strain of L. edodes.
1. Introduction
Mating is an important biological process to generate genetic diversity in nature. In fungi, the mating process is initiated by the physical interaction of mating pheromone with pheromone receptors. The pheromone–receptor interaction activates the mating signal transduction pathway that transfers mating signals to the nucleus of each mating partner [Citation1]. Activation of the mating signal transduction pathway in Saccharomyces cerevisiae occurs via the mitogen-activated protein kinase (MAPK) cascade upon binding of mating pheromone, a- or α-factor, to membrane-embedded pheromone receptor, Ste2 or Ste3, leading to eventual activation of the transcription factor Ste12 which controls the expression of mating-related genes [Citation2,Citation3].
Fungi belonging to Basidiomycota also require the physical interaction between mating pheromone and membrane receptors to initiate the mating process [Citation1]. However, different from S. cerevisiae, the pheromone and receptor genes are genetically linked and are found as pair(s) at a certain chromosomal locus, namely the B mating-type locus [Citation4–6]. The B mating-type locus is further diversified by allelic variation and paralogous expansion/constriction of the pheromone-receptor gene pair [Citation4,Citation5]. Genome sequence analyses on various mushroom species reveal detailed information on the structures of the B mating-type loci. The B mating-type locus of Schizophyllum commune is composed of linked Bα and Bβ subloci in 6.1 kb distance, each of which contains a single pheromone receptor gene (bar3 and bbr2, respectively) plus 16 pheromone genes [Citation4]. S. commune also has 4 non-mating-type receptor genes (brl1–4), brl1–3 in the B mating-type locus, and brl4 in the different scaffold. A recent study has shown their functions in mating (Brl1), hyphal growth (Brl2), and dikaryotic asymmetrical growth (Brl3 and Brl4) [Citation5]. Different from S. commune, PR-a and PR-b subloci in the B mating-type of Flammulina velutipes are located very far from each other (181 kb) [Citation6]. PR-a and PR-b consist of STE3-like receptor gene and one and two pheromone genes, respectively. The B mating-type locus in Coprinopsis cinerea spreads on 35 kb chromosomal region where three subloci, consisting of three variants of STE3-like receptor with 2–4 pheromone genes, are present [Citation7–9].
The B mating-type locus of Lentinula edodes, a popular edible mushroom species in East Asia, is consisted of closely linked Bα and Bβ subloci, each of which contains a pair of pheromone gene and a STE3-type pheromone receptor gene [Citation10]. Our previous investigation on L. edodes has revealed that the B mating-type diversity was generated by combinations of 5 allelic variants of pheromone receptors (RCB1s) from Bα sublocus and 3 variants of pheromone receptors (RCB2s) from Bβ sublocus together with 15 mating pheromones (PHBs), resulting in 15 B mating-types [Citation11]. Similar B mating-type diversities were discovered from C. cinerea (79 B mating-types) and S. commune (81 B mating-types) [Citation7,Citation12–15]. Considering the diversity of the pheromones and receptors, the selective recognition of pheromones by receptors is an important process in determining mating compatibility. The pheromone-receptor specificity of L. edodes was demonstrated by using a yeast model system using synthetic pheromones [Citation16,Citation17]. It was suggested that a certain PHB recognizes a specific partner Rcb from the same sublocus of different B mating-type as a means of non-self recognition in the mating of L. edodes [Citation16,Citation17].
In addition to the B mating-type locus, a new mating pheromone-receptor gene pair was found in L. edodes at a distance of 43.3 kb from the B mating-type locus. The presence of this gene pair has drawn our attention because of its uniqueness among mushroom species. Here, we report detailed gene structures in comparison with PHBs and RCBs from the B mating-type locus. We also report the specificity of the new pheromone receptor to the mating pheromones using the yeast model system, and we propose the biological role for this new gene pair in the regulation of A mating-type gene expression.
2. Materials and methods
2.1. Strains and culture conditions
A total of 26 strains of L. edodes (9 wild and 17 cultivated strains), which were described in our previous study [Citation11], were subjected to the current analysis. L. edodes was cultivated in potato dextrose broth (PDB; Oxoid, Hampshire, UK) or on potato dextrose agar (PDA; Oxoid) at 25 °C. S. cerevisiae RCY1432 (W303a sst2Δ::HIS3, gpaLe::TRP1) [Citation17] was a host strain to construct RCBN-expressing model yeast. The yeasts were grown in yeast extract-peptone-dextrose medium (YPD; 1% yeast extract, 2% peptone, 2% glucose) at 30 °C.
2.2. Sequence analysis
The genomic sequence information of L. edodes B17 [Citation18] was obtained from MycoCosm site (Joint Genome Institute, US Department of Energy). The Bα-N sublocus was identified by BLASTp analysis using L. edodes Rcb1 protein as a query sequence. Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) was employed for the multiple sequence comparison. A phylogenetic tree was constructed using MEGA7 (Maximum Likelihood method, 1000 repeats of bootstrapping) [Citation19]. The transmembrane domain in the mating pheromone receptor protein was predicted by the Phobius program (https://phobius.sbc.su.se/).
2.3. Isolation of the RCBN gene and construction of yeast strains
L. edodes was grown in a PDB medium with gentle shaking for 10 d at 25 °C. The mycelia were collected by filtration with Miracloth (Merck, Darmstadt, Germany). The collected mycelia were ground in liquid nitrogen. The mycelial powder was subjected to total RNA extraction using RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). Reverse transcriptase-PCR (RT-PCR) was performed to generate cDNA for RCBN using a HiPi RT-Prime Kit (ELPIS, Daejeon, Korea). The RCBN amplicon was obtained by PCR using the cDNA and a specific primer set (Supplementary Table S1). The PCR conditions were as follows: hold for 5 min at 95 °C; 25 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 60 s; and 5 min at 72 °C for a final extension.
For the pheromone-RcbN interaction study, S. cerevisiae RCY1467 (W303a, sst2Δ::HIS3, gpaLe::TRP1, ste2Δ::KanMX6RCB-N) was generated by the integration of L. edodes RCBN into the STE2 site of S. cerevisiae RCY1432 (W303a, sst2Δ::HIS3, gpaLe::TRP1). The integration procedure is shown in Supplementary Figure S1. Yeast transformation was performed by standard PEG transformation procedure and the transformant was selected on YPD medium containing G-418 (200 µg/ml; Sigma-Aldrich, St. Louis, MO).
2.4. Interaction of PHBN1 or PHBN2 with RcbN
Two pheromone peptides (PHBN1: EHDSEATADTGFC-OMe and PHBN2: EHTDESGSTADTGFC-OMe) with C-terminal methyl esterification were synthesized through a commercial service (Genscript, Piscataway, NJ). The physical interaction between pheromones and RcbN was monitored through the level of FUS1 gene expression in S. cerevisiae RCY1467. For this, PHBN1 or PHBN2 was treated for 6 h to yeast cells grown in YPD (OD600 = 0.6) at the concentration of 20 µg/ml. After the treatment, the yeast cells were collected by centrifugation at 3,500 rpm for 10 min and the FUS1 gene expression was investigated by qRT-PCR using the previously described procedure [Citation17]. The FUS1 expression was determined using β-tubulin as a reference gene. All experiments were triplicated.
The effect of PHBN1 or PHBN2 on the expression of mating-related genes in L. edodes was also investigated by the treatment of the pheromones in 20 µg/ml to the L. edodes SJ701-M1 strain which was grown in PDB for 10 d. The expression levels of clp1, priA, znf2, HD1, and HD2 were determined as previously described method [Citation17].
3. Results
3.1. Structure analysis of the new mating pheromone and receptor pair
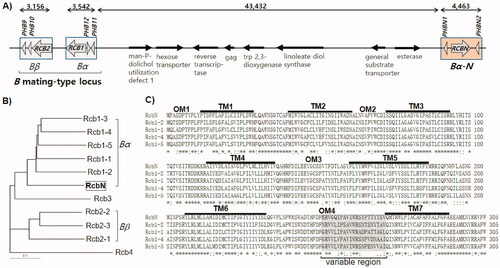
The genome sequence analysis revealed a novel pheromone-receptor gene set 43.3 kb away from the B mating-type locus in the genomic scaffold No. 5 of L. edodes B17 (). The new locus consisted of a STE3-type receptor gene (RCBN) and two pheromone genes (PHBN1 and PHBN2). Eight genes separated the new locus from the B mating-type locus, including mannitol phosphate dolichol utilization defect 1, hexose transporter, and a putative retrotransposon, containing genes for reverse transcriptase (RTase) and gag protein. RcbN, a potential pheromone receptor protein, was homologous to Rcb1 family membrane receptors, which are mating pheromone receptors found from the Bα sublocus of L. edodes (). RcbN was a transmembrane protein containing 7 transmembrane domains (TMs), 4 outer membrane domains (OMs), and 4 cytoplasmic domains (CMs), according to a domain analysis of RcbN and Rcb1 proteins (). Because the other OM domains were substantially homologous to all Rcb1 proteins, a variable region discovered in OM4 (, gray box in OM4) was expected to play a crucial role in the pheromone-specific interaction.
Figure 1. Sequence analysis the Bα-N locus in Lentinula edodes. (A) Genetic structure of the B mating-type locus and the Bα-N locus. The sequence was retrieved from the genome sequence of L. edodes B17 strains (Scaffold #5: 937,000–997,000). The numbers indicate the DNA length in bps; (B) Phylogenetic analysis of the mating pheromone receptor proteins in L. edodes. RcbN in the Bα-N locus is belonged to the Bα family mating pheromone receptors; (C) Topology of the Bα family mating pheromone receptors predicted by Phobius program. Transmembrane domains and outer membrane domains are numbered after TM and OM prefixes, respectively. The variable regions in the OM4 domains are shown in gray box.
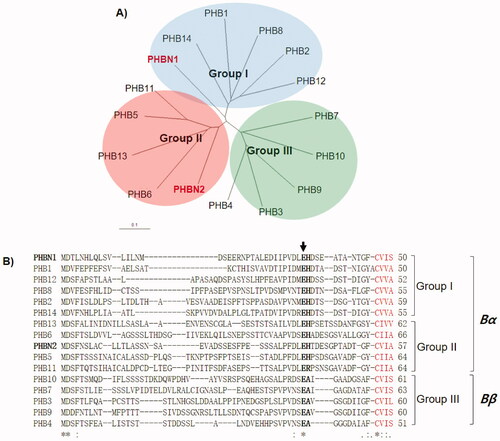
The two pheromones, PHBN1 and PHBN2, belonged to the L. edodes mating pheromone Group I and Group II, respectively, which had previously been referred to as the Bα mating pheromones () [Citation11]. Both pheromones had a CAAX (C, cysteine; A, aliphatic residue; X, any amino acid) motif at the C-terminus (), suggesting that they, like yeast a-factor and L. edodes mating pheromones, are membrane-bound pheromones through the C-term farnecylation [Citation20,Citation21]. Because the organization of Group I and/or Group II pheromones with an RCB1 receptor is a characteristic genetic structure of the Bα sublocus, we named the novel locus as the Bα-N locus.
Figure 2. Characterization of the mating pheromones (PHBs) of Lentinula edodes. (A) Phylogenetic analysis of PHBs from the B mating-type and the Bα-N loci. PHBN1 and PHBN2 are belonged to the mating pheromone Group I and Group II, respectively; (B) Sequence comparison of PHB polypeptides. Maturation of PHBs occurs by proteolytic cleavage (indicated by arrow) and C-terminal modifications at the CaaX motif (highlighted in red).
3.2. Analysis of the Bα-N locus in different strains of L. edodes
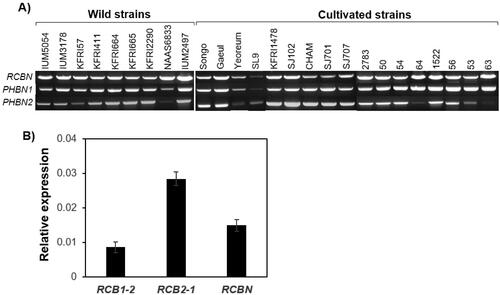
The Bα-N locus in 9 wild strains and 17 cultivated strains of L. edodes was examined using the primer set specific for the Bα-N locus (Supplementary Table S1). Unlike the Bα sublocus, which consists of allelic variants of RCB1 with different sets of mating pheromone genes depending on the mating-type of the strains [Citation11], the Bα-N sequence was nearly identical across all strains, with only a few minor changes (Supplementary Data S1), allowing single primer sets to detect PHBN1, PHBN2, and RCBN (). To verify the functionality of the Bα-N sublocus, expression of RCBN in L. edodes SJ701-M1 strain, which contains RCB1-2 in the Bα sublocus and RCB2-1 in the Bβ sublocus [Citation11], was analyzed by quantitative RT-PCR (qRT-PCR). The qRT-PCR analysis revealed that RCBN was expressed 1.5 fold higher and 0.5 fold lower than RCB1-2 and RCB2-1, respectively (), suggesting that the Bα-N sublocus is functional in L. edodes although whose detailed function is yet to be verified.
Figure 3. Detection of the Bα-N locus in various strains of Lentinula edodes and expression analysis of the mating pheromone receptor genes. (A) PCR detection of RCBN, PHBN1, and PHBN2 in the cultivated and wild strains of L. edodes; (B) qRT-PCR analysis of RCB1-2 (Bα sublocus), RCB2-1 (Bβ sublocus), and RCBN (Bα-N locus) in L. edodes SJ701-M1. The relative expression levels of the mating pheromone receptor genes were calculated by comparing them to the expression amount of β-tubulin. The error bars are standard errors derived from the triplicated experiments.
3.3. Analysis of pheromone–receptor interaction in yeast model system
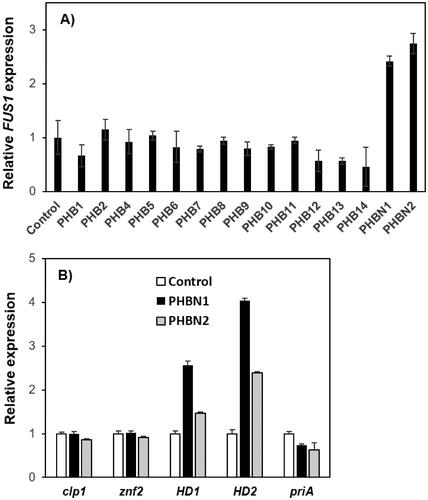
The biological role of Bα-N was assessed through the investigation of RcbN-pheromone interaction with the L. edodes mating pheromones. To this end, a yeast model system was constructed by replacing the S. cerevisiae mating pheromone receptor (STE2) with L. edodes RCBN (Supplementary Figure S1, strain RCY1467), similar to our previous report [Citation17]. The C-term carboxymethylated synthetic pheromones, including the Bα-specific pheromones (PHB1, PHB2, PHB5, PHB6, PHB8, and PHB11–14), the Bβ-specific pheromones (PHB4, PHB7, PHB9, and PHB10), and the Bα-N pheromones (PHBN1 and PHBN2), were treated to actively growing cells of RCY1467 (OD600 = 0.5) for 2 h and then the FUS1 expression was examined as a measure of pheromone-receptor interaction. The yeast cells did not respond to the mating pheromones from the Bα- and Bβ-specific pheromones, as shown in . PHBN1 or PHBN2 treatments, on the other hand, resulted in 2.5- and 2.7-fold increased FUS1 expression, respectively, suggesting the physical interaction between RcbN and PHBN1 or PHBN2 ().
Figure 4. Physical interaction of the mating pheromones with RcbN. (A) PHB-RcbN interaction in yeast model system. Saccharomyces cerevisiae RCY1467, replacing STE2 with Lentinula edodes RCBN, was treated with the B mating-type pheromones (PHB1–14) and the Bα-N pheromones (PHBN1 and PHBN2) in 20 µg/ml. The interaction between PHB and RcbN was determined by the expression of FUS1 gene as a measure of the activation of the yeast mating signal transduction pathway; (B) Expression of the mating-related genes in L. edodes SJ701-M1 upon the treatment of PHBN1 or PHBN2 in 20 µg/ml. The error bars are standard errors derived from the triplicated experiments.
3.4. Mating gene expression in L. edodes by Bα-N pheromones
Since the physical interaction between RcbN and PHBN1/PHBN2 was confirmed by the yeast model system, we investigated the expression of genes known to be responsive to the mating pathway activation in L. edodes. Synthetic PHBN1 or PHBN2 (20 µg/ml) was treated to the culture medium in which the monokaryotic L. edodes SJ701-M1 strain was grown for 3 d. The total RNA extracted from the mycelial cells was subjected to qRT-PCR analysis using specific primer sets targeting clp1, znf2, HD1, HD2, and priA. The qRT-PCR analysis revealed that the mating-related genes were mostly unresponsive upon the treatment of PHBN1 or PHBN2, except for HD1 and HD2 (). HD1 and HD2 are the two genes constituting the A mating-type locus and are known to encode heterodimeric protein complex to control A-related mating process as a transcription factor [Citation22].
4. Discussion
The B mating-type locus of basidiomycetes consists of mating pheromone(s) and pheromone receptors in pairs. The pheromone and receptor pair can occur at a single locus as shown in Pleurotus eryngii [Citation23]. However, the B mating-type locus is normally composed of multiple pheromone and receptor pairs, although the numbers are differed depending on fungal species. S. commune has two subloci at a close distance, each of which contains a receptor and multiple pheromone genes whereas C. cinerea has 3–4 mating-type specific subloci, which are spread on 35 kb chromosomal region [Citation1]. The subloci are not necessarily located at a close distance. The two subloci of F. velutipes B mating-type locus are separated by 181 kb distance [Citation6].
L. edodes has been reported to carry two tightly linked mating pheromone (PHB)-receptor (RCB) gene pairs in the B mating-type locus, constituting the Bα and Bβ subloci [Citation11]. The B mating-type diversity is achieved through allelic variations in RCB1 of Bα and RCB2 of Bβ. Reciprocal interaction between PHB from a mating partner and Rcb from the other partner of the same sublocus can activate the whole mating process [Citation11]. Besides the B mating-type locus, we found a new PHB-RCB pair (PHBN1, PHBN2, and RCBN) at a distance of 43 kb in the same genomic scaffold. Different from the PHBs and RCBs in the B mating-type locus, there was no allelic variation in PHBN1, PHBN2, and RCBN as revealed by sequencing the Bα-Ν sublocus of 26 strains. It appeared that Bα-Ν is an outcome of paralogous expansion of ancestral Bα sublocus, since RcbN1 was homologous to Rcb1 and both of the pheromones belonged to the Bα mating pheromone family. The presence of RTase and gag genes in between the B mating-type locus and the Bα-Ν sublocus implies the possible role of retrotransposon in this gene expansion.
The biological function of the Bα-Ν sublocus was assessed using a yeast model system and synthetic pheromones. RcbN interacts with self-pheromones PHBN1 and PHBN2, rather than pheromones from the B mating-type locus (). The increased expression of two mating-related genes in L. edodes after treatment with PHBN1 and PHBN2 further validated this interaction. Notably, HD1 and HD2, which make up the A mating-type locus, were the two genes that responded to PHBN1 and PHBN2. The A and B mating-type loci are known to be involved in the mating process independently [Citation1], but there are no studies on how the A mating-type locus is activated. In this regard, our findings suggest that Bα-Ν activates the A mating-type locus independently of the B mating pheromone-receptor interaction.
Supplemental Material
Download MS Word (19.2 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Kues U. From two to many: multiple mating-types in basidiomycetes. Fungal Biol Rev. 2015;29:126–166.
- Nakayama N, Miyajima A, Arai K. Nucleotide sequences of ste2 and ste3, cell type-specific sterile genes from Saccharomyces cerevisiae. EMBO J. 1985;4(10):2643–2648.
- Caldwell GA, Naider F, Becker JM. Fungal lipopeptide mating pheromones: a model system for the study of protein prenylation. Microbiol Rev. 1995;59(3):406–422.
- Ohm RA, de Jong JF, Lugones LG, et al. Genome sequence of the model mushroom Schizophyllum commune. Nat Biotechnol. 2010;28(9):957–963.
- Wirth S, Freihorst D, Krause K, et al. What role might non-mating receptors play in Schizophyllum commune? J Fungi. 2021;7(5):399.
- Wang W, Lian L, Xu P, et al. Advances in understanding mating type gene organization in the mushroom-forming fungus Flammulina velutipes. G3. 2016;6(11):3635–3645.
- Riquelme M, Challen MP, Casselton LA, et al. The origin of multiple B mating specificities in Coprinus cinereus. Genetics. 2005;170(3):1105–1119.
- Kues U. Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol Mol Biol Rev. 2000;64:316–353.
- Olesnicky NS, Brown AJ, Honda Y, et al. Self-compatible B mutants in Coprinus with altered pheromone-receptor specificities. Genetics. 2000;156(3):1025–1033.
- Wu L, van Peer A, Song W, et al. Cloning of the Lentinula edodes B mating-type locus and identification of the genetic structure controlling B mating. Gene. 2013;531(2):270–278.
- Ha B, Moon YJ, Song Y, et al. Molecular analysis of B mating-type diversity in Lentinula edodes. Sci Hortic. 2019;243:55–63.
- O’Shea SF, Chaure PT, Halsall JR, et al. A large pheromone and receptor gene complex determines multiple B mating type specificities in Coprinus cinereus. Genetics. 1998;148(3):1081–1090.
- Halsall JR, Milner MJ, Casselton LA. Three subfamilies of pheromone and receptor genes generate multiple B mating specificities in the mushroom Coprinus cinereus. Genetics. 2000;154(3):1115–1123.
- Parag Y, Koltin Y. The structure of the incompatibility factors of Schizophyllum commune: constitution of the three classes of B factors. Mol Gen Genet. 1971;112(1):43–48.
- Raper JR, Krongelb GS, Baxter MG. The number and distribution of incompatibility factors in Schizophyllum commune. Am Nat. 1958;92(865):221–232.
- Ha B, Kim S, Kim M, et al. Activation of the mating pheromone response pathway of Lentinula edodes by synthetic pheromones. Mycobiology. 2018;46(4):407–415.
- Kim S, Ha B, Kim M, et al. Investigation of mating pheromone–pheromone receptor specificity in Lentinula edodes. Genes. 2020;11(5):506.
- Shim D, Park SG, Kim K, et al. Whole genome de novo sequencing and genome annotation of the world popular cultivated edible mushroom, Lentinula edodes. J Biotechnol. 2016;223:24–25.
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874.
- Michaelis S, Barrowman J. Biogenesis of the Saccharomyces cerevisiae pheromone a-factor, from yeast mating to human disease. Microbiol Mol Biol Rev. 2012;76(3):626–651.
- Michaelis S, Herskowitz I. The a-factor pheromone of Saccharomyces cerevisiae is essential for mating. Mol Cell Biol. 1988;8(3):1309–1318.
- Casselton LA, Olesnicky NS. Molecular genetics of mating recognition in basidiomycete fungi. Microbiol Mol Biol Rev. 1998;62(1):55–70.
- Ju Y, Kim S, Kim M, et al. Structure analysis of a and B mating-type loci in a representative commercial strain of Pleurotus eryngii. Sci Horti. 2020;274:109686.
