Abstract
Although Apiospora Sacc. has previously been considered a sexual morph of Arthrinium species on the basis of phylogenetic, morphological, and ecological diagnoses, a recent study delimited these as different species. Recently, 14 species, including eight new species, of marine Arthrinium have been reported from Korea. Six known species have previously been renamed as species in the genus Apiospora (A. arundinis, A. marii, A. piptatheri, A. rasikravindrae, A. sacchari, and A. saccharicola). However, the eight new species of marine Arthrinium (Ar. agari, Ar. arctoscopi, Ar. fermenti, Ar. koreanum, Ar. marinum, Ar. pusillispermum, Ar. sargassi, and Ar. taeanense) are yet to be studied, and thus the taxonomic status of these species remains to be clarified. In this study, we conducted phylogenetic analyses using the internal transcribed spacer, 28S large subunit ribosomal RNA gene, translation elongation factor 1-alpha, and beta-tubulin regions to confirm the phylogenetic position of these eight species. Based on these analyses, we re-identified the eight Arthrinium species as new combinations in Apiospora. Additionally, among the six known Apiospora species, two (A. piptatheri and A. rasikravindrae) have not previously been recorded in Korea. On the basis of morphological and molecular analyses, we report these as new species in Korea. Herein, we present scanning electron micrographs detailing the morphologies of these species, along with phylogenetic trees and detailed descriptions.
1. Introduction
The genus Apiospora Sacc., in the family Apiosporaceae, was recognized and established by Saccardo (1875) with A. montagnei designated as the type species [Citation1]. Historically, several Arthrinium-like species have been identified and synonymized as Arthrinium based on their conidia, conidiophores, and conidiogenous cell shapes [Citation2,Citation3]. Apiospora has also been widely accepted as a synonym for Arthrinium after Ellis [Citation4]. Accordingly, Cordella, Pteroconium, and Scyphospora, considered asexual genera of Apiospora, have also been adopted as synonyms for Arthrinium [Citation5]. Subsequently, Apiospora species were determined to be sexual morphs of Arthrinium species and synonymized under Arthrinium by Crous et al. [Citation6]. Genetic information pertaining to Ar. caricicola, which is regarded as the Arthrinium-type species, was generated along with that for other Arthrinium species (Ar. curvatum and Ar. sporophleum) by Pintos et al. [Citation7]. These species were placed in a monophyletic clade along with Ar. japonicum and Ar. puccioides, which have been phylogenetically distinguished from other Arthrinium species [Citation8]. Morphologically, Arthrinium species differ from Arthrinium s. str species with respect to the ellipsoidal conidia when observed in face view, which appear lenticular in side-view, whereas Arthrinium s. str species are characterized by a diverse range of conidial shapes (cashew-nut, curved, curved with horn-like tips, fusiform, navicular, polygonal, and spherical) [Citation9–11]. Ecologically, Arthrinium have been globally reported (in tropical, subtropical, Mediterranean, temperate, and cold regions) as endophytes, plant pathogens, and saprobes, and are commonly found in a large range of terrestrial environments, including the atmosphere, plants (particularly those in the family Poaceae), soil, and even marine substrates [Citation6,Citation12–15]. In contrast, Arthrinium s. str species have been isolated primarily from plants in the families Cyperaceae and Juncaceae, and are seldom found in tropical and subtropical regions [Citation8]. On the basis of these findings, Pintos et al. [Citation8] reported that Arthrinium and Arthrinium s. str differ genetically, morphologically, and ecologically, and consequently separated these taxonomically. Accordingly, 55 Arthrinium species have been separated from Arthrinium s. str and placed within a new genus, Apiospora. Additionally, on the basis of phylogenetic analyses, Tian et al. [Citation16] reported 13 new combinations and a new Apiospora species. Consequently, a total of 131 Apiospora are currently listed in the Index Fungorum 2021 [Citation17].
Recently, eight new Arthrinium and six known Apiospora species, isolated from marine-derived substrates, have been reported in Korea [Citation12], among which, two known species (A. piptatheri and A. rasikravindrae) have not previously been recorded in Korea. As yet, however, the phylogenetic relationships of the eight new Arthrinium species with the Ar. caricicola clade are yet to be diagnosed. In this study, we have reported the two species A. piptatheri and A. rasikravindrae as new to Korea. We also present detailed morphological descriptions and multi-gene phylogenetic trees constructed based on internal transcribed spacer (ITS), 28S large subunit ribosomal RNA gene (LSU), translation elongation factor 1-alpha gene (TEF), and beta-tubulin (TUB) sequences. Additionally, we performed phylogenetic analyses to establish the taxonomic status of the eight Arthrinium species with respect to members of the Ar. caricicola clade.
2. Materials and methods
2.1. Strains
The cultures of four strains (KUC21220, KUC21279, KUC21327, and KUC21351) were obtained from the Korea University Fungus Collections (KUC). Representative strains have been deposited in the Korean Collection for Type Culture, Daejeon, Korea (KCTC) and the Collection of the National Institute of Biological Resources, Incheon, Korea (NIBR).
2.2. Phylogenetic analysis
Sequences of the four strains were obtained from previous studies [Citation12,Citation18]. For phylogenetic analyses, the ITS, LSU, TEF, and TUB sequences of these strains, along with corresponding Apiospora and Arthrinium reference sequences obtained from GenBank, were assembled, proofread, and edited using MEGA v. 7 [Citation19], and were subsequently aligned using MAFFT 7.130 [Citation20]. ITS, LSU, TEF, and TUB multi-gene phylogenetic analyses were carried out using the maximum likelihood (ML) and Bayesian inference (BI) procedures, as previously described [Citation12]. ML analysis was performed using RAxML v. 7.03 and a GTR + G model with 1000 bootstrap replicates [Citation21], and the BI method was carried out using MrBayes version 3.2, with the best model being selected for each marker [Citation22]. Model tests for ITS, LSU, TEF, and TUB were performed using jModeltest v. 2.1.10 [Citation23]. Markov chain Monte Carlo iterations were run for 5 million generations, and the trees were sampled at successive 1000th generations. Posterior probabilities (PP) were calculated after discarding the initial 25% of sampled trees as burn-in in the majority rule consensus tree. The combined sequence of ITS, TEF, and TUB were analyzed using the same methods and models assigned for each locus.
In the case of Apiospora piptatheri AP4817A (= CBS 145149), information for only ITS and TEF sequences is currently available in GenBank. In addition, the TEF sequence of A. piptatheri AP4817A covers only primer EF2 to primer EF1-1567R [Citation24,Citation25], which does not overlap with the majority of the Apiospora TEF sequences examined in this study. Thus, the reference sequences for A. piptatheri AP4817A that contain information corresponding to that of the available TEF sequence were individually recollected from GenBank and aligned using MAFFT 7.130 [Citation20]. ITS sequences of the reference strains were also obtained from GenBank. Assessments of the BI models for ITS and TEF were performed individually. The combined sequences of ITS and TEF from A. piptatheri AP4817A were analyzed using the same methods. All analyses were performed using CIPRES [Citation26], and the trees thus constructed were visualized using FigTree v 1.4.3 [Citation27] and edited using Adobe Illustrator CS6.
2.3. Morphological studies
Morphological studies were performed by culturing strains on oatmeal agar (OA: 7.25% Difco Oatmeal Agar; Difco, Detroit, MI, USA), potato dextrose agar (PDA: 3.9% Difco Potato Dextrose Agar; Difco), and malt extract agar (MEA: 2% Bacto malt extract/1.5% Bacto Agar; Difco). Culture characteristics, including growth rates, surface structures, aerial mycelium, colony shape and color, and sporulation, were recorded. A Munsell color chart was used to apply a standard color code [Citation28]. To determine fungal growth rates, colony diameters were measured in the dark at 15 °C, 20 °C, and 25 °C at 24-h intervals for 14 days. Culture colonies were mounted in water for microscopic examination of the conidia, conidiogenous cells, and hyphae using an Olympus BX51 light microscope (Olympus, Tokyo, Japan) fitted with a DP20 microscope camera (Olympus). Values for each of the examined morphological characters were based on at least 30 independent measurements. Scanning electron microscopy was performed commercially by Wooyoung Solution Inc. (Suwon, Korea) using an S-5200 scanning electron microscope (Hitachi, Tokyo, Japan).
3. Results
3.1. Phylogenetic analysis
Multi-gene phylogenetic analyses were performed using the ML and BI methods. The multilocus datasets contained 152 taxa, including those of Melogramma campylosporum MBU (Melogrammataceae), Allelochaeta acuta CBS 144168 (Sporocadaceae), and Nigrospora gorlenkoana CBS 480.73 (Trichosphaeriacea) used as outgroups, consisting of 4418 characters, including gaps (). In the BI analysis, ITS and TUB sequence datasets were assigned as HKY + I + G to the best-fit model, and LSU and TEF sequence datasets were assigned as GTR + I + G and GTR + G, respectively, to the best-fit model. Topologies of the ML and BI trees were found to be identical, and thus we selected the BI tree to be representative (). Within the constructed tree, the eight species of marine Arthrinium (see blue text in ) were found to cluster in the Apiospora clade, and the four strains KUC21220, KUC21279, KUC21327, and KUC21351 were identified as belonging to the A. piptatheri or A. rasikravindrae clades (see boldface red text in ). KUC21220 and KUC21279 formed a monophyletic group with A. piptatheri CBS 145149 with a high support value (). Although KUC21327 and KUC21351 showed low resolution in the multigene phylogeny (), they can be well distinguished with high support (ML bootstrap support = 92%, Bayesian posterior probabilities = 0.96) from other Apiospora species in the combined ITS, TEF, and TUB tree (Supplemental Figure 1S).
Figure 1. A Bayesian tree based on a concatenated alignment of ITS, LSU, TEF, and TUB sequences. The node numbers indicate the ML bootstrap support (BS) > 70% and Bayesian posterior probabilities (PP) > 0.70 as BS/PP. BS and PP values of less than 70% and 0.70, respectively, are indicated by hyphens (“-”). The fungal cultures examined in this study are shown in boldface red type. The eight new combinations are denoted by a blue color.

3.2. Taxonomy
Apiospora piptatheri (Pintos & P. Alvarado) Pintos & P. Alvarado, Fungal Systematics and Evolution 7: 207 (2021) [MB#837708] ().
Figure 2. Apiospora piptatheri KUC21220. (a) Colonies on PDA, (b) MEA, and (c) OA (top). (d) Colonies on PDA, (e) MEA, and (f) OA (f) (low), (g–h) conidia under SEM, (i–k) conidia attached conidiogenous cells. Scale bar = 10 μm.
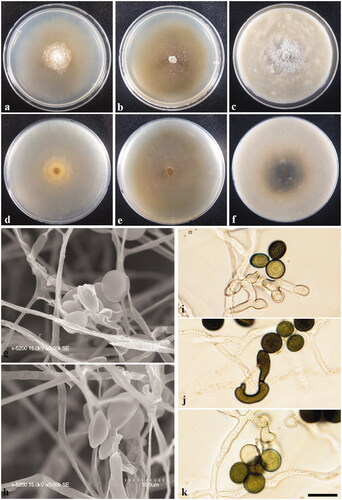
Colony diameters – measured at 120 h (mm): 15 °C: PDA 6–9, MEA 11–13, and OA 11–12; 20 °C: PDA 12–13, MEA 21–27, and OA 15–17; 25 °C: PDA 6–9, MEA 8–12, and OA 9–11.
Culture characteristics – PDA, colonies low, concentrically spreading with sparse aerial mycelium, margin circular; mycelia yellowish ivory colored; sporulation on hyphae after 2 weeks, spreading around a center, spore black; pigment absent; no distinctive odor. MEA, colonies low, flat, concentrically spreading, thin with aerial mycelium, margin circular; mycelia white colored, sparsely reverse olive; sporulation on hyphae around a center after 2 weeks, spore black; pigment absent; no distinctive odor. OA, colonies abundant, concentrically spreading, dense at center with aerial mycelium, margin circular; mycelia white to greenish gray colored, usually reverse green from center; sporulation on hyphae, spore black; dark green (5GY 2.5/1) pigment diffused from center in media and yellow pigment diffused around center; no distinctive odor.
Asexual morphology – Conidiophore reduced to conidiogenous cells. Conidiogenous cells at first pale green, becoming brown, polyblastic, sympodial, cylindrical to ampulliform. Conidia green to brown, smooth, globose to ellipsoid, (7.3–)7.6–9.0(–9.7) × (6.7–)6.8–8.9 μm; lenticular in side-view, with equatorial slit, 4.5–6.1 μm wide, elongated cell observed. Mycelium consisting of smooth, hyaline, branched, septate.
Specimen examined. SOUTH KOREA, Jeju-do, 33°23′39.2″N, 126°14′23.0″E, isolated from Sargassum fulvellum, 10 Jan. 2015, S. Jang (KUC21220 and KUC21279).
Remarks – The culture characters of A. piptatheri KUC21220 grown on MEA medium are similar to those of the original description (colonies flat, spreading, aerial mycelium) [Citation7]. The diameters of the conidial cells of A. piptatheri (KUC21220 and KUC21279) were found to be wider than those of the ex-type culture of A. piptatheri [CBS 145149 = AP4817A, globose to ellipsoid, 6–8 × 3–5 μm (n = 30)] (Supplemental Table S2) [Citation7]. However, the phylogenetic analyses provided strong evidence to indicate that these are the same species (, Supplemental Figure S2). This, to the best of our knowledge, is the first report of A. piptatheri from a marine environment in South Korea, for which we provide a detailed characterization of cultures grown on PDA, MEA, and OA media.
Apiospora rasikravindrae (Shiv M. Singh, L.S. Yadav, P.N. Singh, Rahul Sharma & S.K. Singh) Pintos & P. Alvarado, Fungal Systematics and Evolution 7: 207 (2021) [MB#837716] ().
Figure 3. Apiospora rasikravindrae KUC21327. (a) Colonies on PDA, (b) MEA, and (c) OA (top). (d) Colonies on PDA, (e) MEA, and (f) OA (low), (g–h), conidia under SEM, (i–k) conidia attached conidiogenous cells. Scale bar = 10 μm.
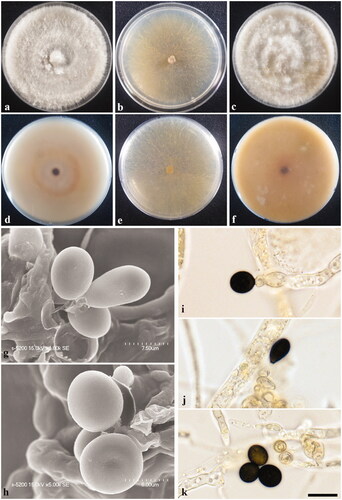
Colony diameters – measured at 120 h (mm): 15 °C: PDA 21–23, MEA 14–17, and OA 12–13; 20 °C: PDA 34–39, MEA 30–35, and OA 28–33; 25 °C: PDA 30–32, MEA 22–25, and OA 29–35.
Culture characteristics – PDA, colonies abundant, concentrically spreading, thick with aerial mycelium, margin circular; mycelia creamy white to pale yellow colored; sporulation on hyphae within 2 weeks, spreading around center, very plentiful, spore black; yellow (10YR 7/6) pigment diffused in media; no distinctive odor. MEA, colonies abundant, flat, concentrically spreading with sparse aerial mycelium, margin irregular; mycelia white to pale yellow colored; sporulation on hyphae around center within 2 weeks, spore black; pigment absent; no distinctive odor. OA, colonies abundant, concentrically spreading, thick and dense at center with aerial mycelium, margin irregular; mycelia creamy white colored; sporulation on hyphae, spore black; yellow (2.5Y 8/8) pigment to brownish yellow (10YR 6/8) pigment diffused in media; no distinctive odor.
Asexual morphology – Conidiogenous locus aggregated in clusters on hyphae; conidiogenous cell at first pale green, becoming green to brown, monoblastic or polyblastic, ampulliform. Conidia brown, smooth, globose to ellipsoid in surface view, (6.9–)7.5–8.8(–9.4) × (6.5–)7.0–8.1(–9.2) μm; lenticular in side-view, with equatorial slit, 4.8–6.5 μm wide. Mycelium consisting of smooth, hyaline, branched, septate, 3.1–4.9 μm diam hyphae.
Specimen examined. SOUTH KOREA, Gangwon-do, Goseong-gun, 38°28′44.0″N, 128°26′18.9″E, isolated from Egg masses of Arctoscopus japonicus, November 10 2016, M.S. Park (KUC21327 and KUC21351).
Remarks – Notably, the diameters of A. rasikravindrae (KUC21327 and KUC21351) conidial cells are smaller than those of the A. rasikravindrae-type culture [NFCCI 2144, lenticular, ovoid in face view, 10–15 × 6.0–10.5 µm (n = 50); elongate to clavate conidia, 15–25 × 7.5–10 µm (n = 50)] (Supplemental Table S2) [Citation29]. However, the conidiophores described in the original description of this species were not observed in this strain. The optimum temperatures for the growth of Apiospora rasikravindrae have been established as 18 °C for an Arctic isolate and 25 °C for Japanese and Chinese strains [Citation29]. Growth of the strains examined in the present study appeared to be optimal within a range of 20–25 °C, which is consistent with those of the type strains. The ITS, LSU, TEF, and TUB sequences of our strains KUC21327 and KUC21351 showed high similarities with those of Apiospora rasikravindrae NFCCI 2144 (ITS: 3/556 (Diff/Total), 99.46% (similarity), LC 5449 (ITS: 2/559, 99.64%; LSU: 0/498, 100%; TEF: 2/433, 99.54%; TUB: 0/394, 100%), LC 7115 (ITS: 1/559, 99.82%; LSU: 2/483, 99.59%; TEF: 2/433, 99.54%; TUB: 1/395, 99.75%), CBS 337.61 (ITS: 1/559, 99.82%; LSU: 2/498, 99.6%), CPC 21602 (ITS: 1/548, 99.82%). On the basis of these results, we identified KUC21327 and KUC21351 as strains of A. rasikravindrae. This is the first report of A. rasikravindrae isolated in South Korea and these are first isolates from a marine environment, for which detailed descriptions and phylogenies have been provided.
3.3. New combinations
Note: The asexual morphologies and ecological characteristics of the eight new combinations are presented in . These eight species of marine Arthrinium have previously been described by Kwon et al. [Citation12]. The spores are similar in shape (globose, subglobose, ellipsoid, elongate ellipsoid in surface view; lenticular in side-view) although differ in size, and their shape appears to be similar to that of spores in species of the genus Apiospora [Citation12]. The eight species were isolated from marine substrates in Korea, notably the eggs of Arctoscopus japonicus and macroalgae such as Agarum cribrosum and Sargassum fulvellum [Citation12]. Phylogenetically, the eight species are well clustered with members of the Apiospora clade (). On the basis of morphological, ecological, and phylogenetic analyses, we thus propose the transfer of these eight marine Arthrinium under new combinations of Apiospora species.
Table 1. Morphological characteristics of conidia and isolation information for the new combinations.
Apiospora agari (S.L. Kwon, S. Jang & J.J. Kim) S.L. Kwon & J.J. Kim, comb. nov.
MycoBank MB834592.
Basionym: Arthrinium agari S.L. Kwon, S. Jang & J.J. Kim., IMA Fungus 12:13. 2021.
Apiospora arctoscopi (S.L. Kwon, S. Jang & J.J. Kim) S.L. Kwon & J.J. Kim, comb. nov.
MycoBank MB834593.
Basionym: Arthrinium arctoscopi S.L. Kwon, S. Jang & J.J. Kim., IMA Fungus 12:13. 2021.
Apiospora fermenti (S.L. Kwon, S. Jang & J.J. Kim) S.L. Kwon & J.J. Kim, comb. nov.
MycoBank MB834594.
Basionym: Arthrinium fermenti S.L. Kwon, S. Jang & J.J. Kim., IMA Fungus 12:13. 2021.
Apiospora koreana (S.L. Kwon, S. Jang & J.J. Kim) S.L. Kwon & J.J. Kim, comb. nov.
MycoBank MB834596.
Basionym: Arthrinium koreanum S.L. Kwon, S. Jang & J.J. Kim., IMA Fungus 12:13. 2021.
Apiospora marina (S.L. Kwon, S. Jang & J.J. Kim) S.L. Kwon & J.J. Kim, comb. nov.
MycoBank MB834595.
Basionym: Arthrinium marinum S.L. Kwon, S. Jang & J.J. Kim., IMA Fungus 12:13. 2021.
Apiospora pusillisperma (S.L. Kwon, S. Jang & J.J. Kim) S.L. Kwon & J.J. Kim, comb. nov. MycoBank MB834597.
Basionym: Arthrinium pusillispermum S.L. Kwon, S. Jang & J.J. Kim., IMA Fungus 12:13. 2021.
Apiospora sargassi (S.L. Kwon, S. Jang & J.J. Kim) S.L. Kwon & J.J. Kim, comb. nov.
MycoBank MB834598.
Basionym: Arthrinium sargassi S.L. Kwon, S. Jang & J.J. Kim., IMA Fungus 12:13. 2021.
Apiospora taeanensis (S.L. Kwon, S. Jang & J.J. Kim) S.L. Kwon & J.J. Kim, comb. nov.
MycoBank MB834599.
Basionym: Arthrinium taeanense S.L. Kwon, S. Jang & J.J. Kim., IMA Fungus 12:13. 2021.
4. Discussion
In this study, two Apiospora species (A. piptatheri and A. rasikravindrae) are reported as previously unrecorded species in Korea, based on morphological and phylogenetic analyses. Moreover, eight new combinations are proposed based on phylogenetic analyses with morphological and ecological diagnoses. The strains KUC21327 and KUC21351 are characterized by conidial cell sizes that differ slightly from those in the original descriptions [Citation29]. However, both strains were well clustered with A. rasikravindrae with high support values in the phylogenetic tree based on a combination of ITS, TEF, and TUB sequences (Supplemental Figure S1). Furthermore, strong sequence homologies and the optimal growth rates of these strains are consistent with ex-type strains, thereby providing support for the proposed taxonomic assignment. Similarly, we established that KUC21220 and KUC21279 were well clustered with A. piptatheri CBS 145149 in the multigene phylogenetic tree (). Moreover, the morphological characters of these strains are generally consistent with those presented in original descriptions. However, given the lack of an A. piptatheri CBS 145149 reference sequence for TUB and the noncorrespondence among the existing partial sequences of TEF, additional analyses will be necessary to confirm the resolution of the multi-gene phylogenetic results. Given that the TEF partial sequence of A. piptatheri CBS 145149 only contained primer EF2 to primer EF1-1567R site, which are not covered in the reference sequences of most Apiospora species, we obtained additional reference sequences covering the sites of TEF and performed ML and BI phylogenetic analyses using combined ITS and TEF datasets. The phylogenetic trees thus obtained revealed that KUC21220 and KUC21279 form a monophyletic group with A. piptatheri CBS 145149 with strong support values (Supplemental Figure S2).
The multigene tree showed a sufficiently high resolution for most Apiospora and Arthrinium species (), which thus enabled us to distinguish Apiospora and Arthrinium clades. However, the two Arthrinium species Ar. trachycarpum (CFCC 53038, CFCC 53039) and Ar. urticae (IMI 326344) were found to be exceptions in this regard, in that they formed a distinct clade separate from those of Apiospora and Arthrinium species, which was positioned close to Allelochaeta acuta CBS 144168 (Sporocadaceae) (). Nevertheless, despite this apparent phylogenetic separation, these two species are characterized by morphologies similar to those of Apiospora or Arthrinium species. For example, Ar. trachycarpum was found to be morphologically similar to species of Apiospora (conidiophores reduced to conidiogenous cells, subglobose to lenticular conidia), whereas Ar. urticae is characterized by morphological traits similar to those of Arthrinium species (conidiophores cylindrical with thick blackish septa, subspherical conidia). In both cases, however, these species differ significantly from Allelochaeta species in terms of morphology [Citation4,Citation30,Citation31]. These findings are consistent with those reported previously [Citation16], thereby highlighting the need for further genetic and morphological analyses to confirm the taxonomic status of these two species.
tmyb_a_2038857_sm9402.docx
Download MS Word (64 MB)tmyb_a_2038857_sm9365.docx
Download MS Word (36.2 KB)tmyb_a_2038857_sm9714.jpg
Download JPEG Image (16.5 MB)tmyb_a_2038857_sm0085.jpg
Download JPEG Image (61.1 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Saccardo P. Conspectus generum pyrenomycetum italicorum additis speciebus fungorum Venetorum novis vel criticis, systemate carpologico dispositorum. Atti della Società Veneziana-Trentina-Istriana di. Scienze Naturali. 1875;4:77–100.
- Hughes SJ. Conidiophores, conidia, and classification. Can J Bot. 1953;31(5):577–659.
- Minter DW. A re-appraisal of the relationships between Arthrinium and other hyphomycetes. Proc: Plant Sci. 1985;94(2–3):281.
- Ellis MB. Dematiaceous hyphomycetes VI. Mycol Pap. 1965;103:1–46.
- Samuels G, McKenzie E, Buchanan DE. Ascomycetes of New Zealand 3. Two new species of Apiospora and their Arthrinium anamorphs on bamboo. NZ J Bot. 1981;19(2):137–149.
- Crous PW, Groenewald JZ. A phylogenetic re-evaluation of Arthrinium. IMA Fungus. 2013;4(1):133–154.
- Pintos Á, Alvarado P, Planas J, et al. Six new species of Arthrinium from Europe and notes about A. caricicola and other species found in Carex spp. hosts. MycoKeys. 2019;49:15–48.
- Pintos Á, Alvarado P. Phylogenetic delimitation of Apiospora and Arthrinium. Fungal Syst Evol. 2021;7:197–221.
- Ellis MB. Dematiaceous hyphomycetes XI. Mycol Pap. 1972;131:1–25.
- Rambelli A, Venturella G, Ciccarone C. More dematiaceous hyphomycetes from Pantelleria mediterranea maquis litter. Flora Mediterranea. 2008;19:81–113.
- Cooke WB. The genus Arthrinium. Mycologia. 1954;46(6):815–822.
- Kwon SL, Park MS, Jang S, et al. The genus Arthrinium (Ascomycota, Sordariomycetes, Apiosporaceae) from marine habitats from Korea, with eight new species. IMA Fungus. 2021;12(1):1–26.
- Wang M, Tan X-M, Liu F, et al. Eight new Arthrinium species from China. MycoKeys. 2018;(34):1–24.
- Jiang N, Liang YM, Tian CM. A novel bambusicolous fungus from China, Arthrinium chinense (Xylariales). Sydowia. 2020;72:77–83.
- Feng Y, Liu J-KJ, Lin C-G, et al. Additions to the genus Arthrinium (Apiosporaceae) from bamboos in China. Front Microbiol. 2021;12:661281.
- Tian X, Karunarathna SC, Mapook A, et al. One new species and two new host records of Apiospora from bamboo and maize in Northern Thailand with thirteen new combinations. Life. 2021;11(10):1071.
- Index Fungorum 2021. Available from: http://www.indexfungorum.org/Names/Names.asp
- Hong J-H, Jang S, Heo YM, et al. Investigation of marine-derived fungal diversity and their exploitable biological activities. Mar Drugs. 2015;13(7):4137–4155.
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874.
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780.
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690.
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–1574.
- Darriba D, Taboada G, Doallo R, et al. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9(8):772.
- O'Donnell K, Sutton DA, Rinaldi MG, et al. Novel multilocus sequence typing scheme reveals high genetic diversity of human pathogenic members of the Fusarium incarnatum-F. equiseti and F. chlamydosporum species complexes within the United States. J Clin Microbiol. 2009;47(12):3851–3861.
- Rehner SA, Buckley E. A Beauveria phylogeny inferred from nuclear ITS and EF1-alpha sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 2005;97(1):84–98.
- Miller M, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE); 2010 Nov; New Orleans, LA.
- Rambaut A. FigTree-version 1.4. 3, a graphical viewer of phylogenetic trees. Computer program distributed by the author. Available from: http://treebioedacuk/software/figtree
- Munsell. Munsell soil-color charts with genuine Munsell color chips. Grand Rapids (MI): Munsell Color; 2009.
- Singh SM, Yadav LS, Singh PN, et al. Arthrinium rasikravindrii sp. nov. from Svalbard, Norway. Mycotaxon. 2013;122(1):449–460.
- Yan H, Jiang N, Liang L-Y, et al. Arthrinium trachycarpum sp. nov. from Trachycarpus fortunei in China. Phytotaxa. 2019;400(3):203–210.
- Liu F, Bonthond G, Groenewald J, et al. Sporocadaceae, a family of coelomycetous fungi with appendage-bearing conidia. Stud Mycol. 2019;92:287–415.
