Abstract
Rhytisma lonicericola was identified as a tar spot fungus on Lonicera sp. in 1902, and has since been recorded on several species of Lonicera in China, Japan, and Korea. Most of the previous records of R. lonicericola have been based on a list of disease occurrences in the absence of any formal morphological identification or molecular analyses. Using six newly obtained specimens collected in the past 2 years, we confirmed the tar spot fungus found on L. japonica in Korea as R. lonicericola based on morphological examinations and molecular phylogenetic analyses. This fungus was distinguished from R. xylostei, another tar spot fungus on Lonicera, by ascospore size and geographical distributions. We present detailed mycological information and, for the first time, DNA sequence data useful for the identification of R. lonicericola.
The genus Rhytisma Fr. belonging to the fungal phylum Ascomycota contains more than 100 taxa (http://www.indexfungorum.org), many of which cause multiple tar spot lesions on plant leaves. The host range of Rhytisma encompasses more than 30 plant genera, including Acer, Andromeda, Aster, Ilex, Lonicera, Lyonia, Rhododendron, and Vaccinium. On species of Lonicera, two Rhytisma species have been reported as tar spot fungi, namely, R. lonicericola Henn., which has been recorded on more than 12 Lonicera species, and R. xylostei Naumov, which has been found only on Lonicera nummulariifolia [Citation1]. Rhytisma lonicericola was first described on an unknown species of Lonicera from Japan in 1902 [Citation2], and since then, it has subsequently been reported to infect other Lonicera spp. in Japan, China, and Korea [Citation1].
In Korea, Lonicera japonica Thunb. (Caprifoliaceae) has been widely planted as an effective groundcover and fragrant ornamental, and its leaves and flowers have also been used as an important source of traditional medicine [Citation3,Citation4]. The tar spot of L. japonica and the associated fungus R. lonicericola were first reported in Korea in 1994 [Citation5]. However, although there are approximately 30 species of Lonicera found growing in Korea, to date, tar-spot infection has only been detected on L. japonica [Citation5,Citation6].
Nevertheless, in both Korea and other countries, most of the previous records of R. lonicericola have been based on a list of disease occurrence, in the absence of any formal morphological or molecular identification of the fungus, which can presumably be attributed to the fact that the large black stromatal lesions are a typical diagnostic feature of tar spots. Accordingly, in this study, we sought to confirm the identity of the tar-spot fungus found on L. japonica in Korea, based on morphological characteristics and molecular phylogenetic analyses.
During recent extensive forays for phytopathogenic fungi, we found typical tar spots on the leaves of L. japonica at Wanju in 2020, Hongcheon in 2020 and 2021, and Wonju in 2021 (). Voucher specimens of the infected tissues were deposited in the Korea University Herbarium under the following collection numbers: KUS-F31821 (July 2 2020; Wanju), F32085 (October 22 2020; Hongcheon), F32170 (November 28 2020; ibid), F32193 (May 6 2021; ibid), F32277 (June 21 2021; ibid), and F32288 (June 24 2021; Wonju).
Figure 1. Tar spots of Lonicera japonica associated with Rhytisma lonicericola. (a, b) Tar spots on the leaf surface in autumn. (c) Small newly infected tar spots in spring. (d) Prominent tar spots on overwintered leaves in May. Note the disease-free, newly expanded leaves. (e) Tar spots on overwintered leaf surface. (f) Opened ascostroma on the leaf surface, exposing the hymenia. (g) Ascomata in vertical section. ads = adaxial leaf surface, ol = outer layer of the stroma, hy = hymenium. (h) Hymenial layer of an ascoma. as = ascus, pa = paraphysis. (i, j) Asci with ascospores. (k, l) Ascospores.

Mature asci and ascospores were observed in the ascomata on the overwintered leaves from late April to early June in our samples (), whereas asci and ascospores were not detected in the two samples collected in late autumn (F32085) and early winter (F32170). During spring, new infections were noticed in early June, and small tar spots had become prominent by late June (). Based on these field observations in Hongcheon (37°40′36′′N 127°52′55′′E) and Wonju (37°21′34′′N 127°53′09′′E), we believe that ascospores mature during spring and act as the initial source of inocula infecting new leaves.
For morphological observations, symptomatic leaves with tar-spot infection were immersed in sterile distilled water for 1 h to promote apothecial opening. To observe the ascomata, the leaves were vertically sectioned to a thickness of 10 μm using a Leica CM 3505 S cryomicrotome (Leica Biosystems, Nussloch, Germany) and the sections were mounted in water. The morphologies of the stromata, ascomata, asci, paraphyses, and ascospores were examined using compound light microscopes (Olympus BX51; Olympus, Tokyo, Japan), and Zeiss AX10 equipped with an AxioCam MRc5 camera; Carl Zeiss, Göttingen, Germany).
The black stromata on the leaf surface were observed to be irregular and amphigenous, with the upper parts being considerably larger than the lower parts (). The stromata were often confluent and granular, with diameters ranging from 1 to 5 mm (). The ascomata, which develop on the adaxial surface of affected leaves, were irregular in shape and located in the stromatal surface when mature (). They opened via a more or less irregular split, exposing a yellow or ivory hymenium on the surface (). In the median vertical sections, the outer layers of the stromata adaxial surface grew to a thickness of 25–40 μm (n = 20) and internally contained tightly packed hyphae and thick-walled hyaline paraphyses (). Ascomata were confined to the adaxial part of the stromata and were 110–175 μm deep with one to two (occasionally three) loculi (n = 20) (). The paraphyses (90–300 × 1–2.5 μm) were filiform, branched, septate, circinately coiled, and swollen at the apex (n = 30) (). The asci (84–152 × 10–15 μm) were 8-spored, narrowly clavate, short-stalked, and rounded or slightly rostrate at the apex (n = 30) (), whereas the ascospores (22–34 × 3.4–5 μm) were hyaline, elongated fusiform, or tapering toward the base with gelatinous sheaths, aseptate, or rarely one-septate (n = 30) (). Some Rhytisma species are known to have a spermogonial stage, although no spermatia were observed in our samples.
To confirm the morphology-based identification, we performed molecular phylogenetic analyses using ITS and LSU sequences. Genomic DNA was extracted from the ascomata growing on two specimens (KUS-F31821 and F32170), using Quick-DNA Fungal/Bacterial Miniprep Kit (Zymo Research, Irvine, CA, USA). The internal transcribed spacer (ITS) and 28S large-subunit (LSU) regions of the fungal rDNA were amplified using the primer pairs ITS1F/ITS4 [Citation7] and LR0R/LR5 [Citation8], respectively. Purified PCR products were sequenced by a DNA sequencing service (Macrogen, Seoul, Korea) with the same primers used for amplification. The generated DNA sequences were analyzed in SeqMan v. 7.1 from the Lasergene package (DNASTAR, Inc. Madison, WI, USA) to obtain consensus sequences. The assembled sequences were compared with entries in the NCBI database using the BLASTn program. For phylogenetic analyses, both ITS and LSU sequences were aligned and manually edited using the corresponding sequences of Rhytisma species retrieved from GenBank. We used Lophodermium piceae as an outgroup taxon (GenBank Accession Nos. KX009052 and HM140551). Two phylogenetic trees were constructed for the ITS and LSU regions using MEGA7 software [Citation9] based on the maximum likelihood (ML) method. The gaps in the aligned sequences were treated as deletions. The strength of the internal branches of the resulting trees was assessed based on a bootstrap analysis of 1000 replicates.
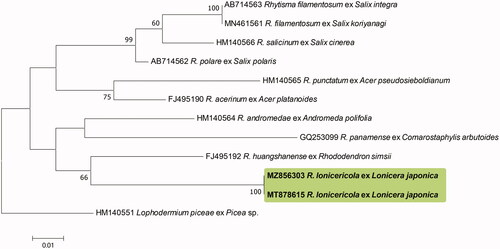
The ITS sequences obtained from the two Korean specimens were found to be 100% identical and formed a monophyletic clade in the tree. The highest sequence similarity (88%) with the other accessions was that of the Korean isolates (GenBank accession nos. MT878614, and MZ856302) and R. huangshanense (GQ253101). There were no Rhytisma spp. in the GenBank database with a sequence similarity greater than 90%. The LSU sequences of the Korean specimens (MT878615 and MZ856303) were also identical and showed the highest similarity (92.73%) with that of R. huangshanense (FJ495192) based on a BLASTn search. Our phylogenetic analyses indicated that the Korean specimens formed a separate clade distinct from the other Rhytisma species used for the ML tree analyses ( and ). Unfortunately, owing to the unavailability of the requisite sequence data in the GenBank database, we were unable to include R. lonicericola in the phylogenetic analyses, which we found to be morphologically closest to the two Korean specimens.
Figure 2. Phylogenetic relationship between Rhytisma lonicericola specimens and the reference isolates of related Rhytisma species retrieved from the GenBank database, inferred using Maximum likelihood method based on an analysis of ITS rDNA region sequences. Bootstrap values based on 1000 replications are indicated at the branches. The scale bar represents 0.02 nucleotide substitutions per site. The Korean specimens characterized in this study are shown in bold.

Figure 3. Phylogenetic relationship between Rhytisma lonicericola specimens and the reference isolates of related Rhytisma species retrieved from the GenBank database, inferred using Maximum likelihood method based on an analysis of LSU rDNA region sequences. Bootstrap values based on 1000 replications are indicated at the branches. The scale bar represents 0.01 nucleotide substitutions per site. The Korean specimens presented in this study are shown in bold.
To date, two Rhytisma species have been reported as tar spot fungi infecting Lonicera spp., namely, R. lonicericola and R. xylostei [Citation1]. Of these, the morphological characteristics of the fungi examined in the present study are most consistent with the original description of R. lonicericola on Lonicera sp. by Henning in 1902 [Citation2] and the previous description by Shin (1994) in Korea [Citation5]. Since its first description in 1914, R. xylostei has been recorded on L. nummulariifolia in Uzbekistan and on Lonicera sp. in Siberia [Citation6,Citation11]. Although the morphological characteristics of the stromata and paraphyses of our fungi are similar to those of R. xylostei, the ascospores are approximately 1.5–2 times longer. Furthermore, the aforementioned two Rhytisma species also have distinct geographical distributions, with R. lonicericola being found mainly in northeast Asia, including China, Japan, and Korea, whereas R. xylostei tends to occur from Central Asia to Siberia [Citation1,Citation6,Citation10,Citation11].
Thus, the findings of this study confirm that the Rhytisma fungus found on L. japonica in Korea is R. lonicericola, and we present additional information relating to the symptomatic and morphological features of R. lonicericola, along with sequence data for two rDNA regions of this fungus.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Farr DF, Rossman AY. Fungal Databases, U.S. National Fungus Collections, ARS, USDA. http://nt.ars-grin.gov/fungaldatabases/
- Engler A. Saccardo’s Syll. Fung. XVIII: 164. Bot Jb German. 1902;32:43.
- Starr F, Starr K, Loope L. Lonicera japonica. Caprifoliaceae. Maui: United States Geological Survey-Biological Resources Division; 2003.
- Shang X, Pan H, Li M, et al. Lonicera japonica Thunb.: ethnopharmacology, phytochemistry and pharmacology of an important traditional chinese medicine. J Ethnopharmacol. 2011;138(1):1–21.
- Shin HD. New fungal diseases of economic resource plants in Korea (І). Korean J Plant Pathol. 1994;10:181–191.
- Engler A. Saccardo’s Syll. Fung. XXIV: 1266. Bull Soc Mycol Fr. 1914;30:425.
- White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocol. 1990;18:315–322.
- Moncalvo JM, Lutzoni FM, Rehner SA, et al. Phylogenetic relationships of agaric fungi based on nuclear large subunit ribosomal DNA sequences. Syst Biol. 2000;49(2):278–305.
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874.
- Mueller E. On the geographical distribution of some parasitic ascomycetes on Lonicera. Advancing frontiers of mycology and plant pathology. In: Bilgrami KS, Misra RS, Misra PC eds. New Delhi: Today & Tommorrows’ Printers & Publishers; 1981. pp. 11–17.
- Kuz’michev EP, Sokolova ES, Kulikova EG, et al. Common fungal diseases of Russian forests. U.S. Department of Agriculture, Forest Service, Northeastern Research Station. 2001. p. 137.
