Abstract
The rare edible and medicinal fungus Antrodia cinnamomea has a substantial potential for development. In this study, Illumina HiSeq 2000 was used to sequence its transcriptome. The results were assembled de novo, and 66,589 unigenes with an N50 of 4413 bp were obtained. Compared with public databases, 6,061, 3,257, and 2,807 unigenes were annotated to the Non-Redundant, Gene Ontology, and Kyoto Encyclopedia of Genes and Genomes databases, respectively. The genes related to terpene biosynthesis in the mycelia of A. cinnamomea were analyzed, and acetyl CoA synthase (ACS2 and ACS4), hydroxymethylglutaryl CoA reductase (HMGR), farnesyl transferase (FTase), and squalene synthase (SQS) were found to be upregulated in XZJ (twig of C. camphora) and NZJ (twig of C. kanehirae). Moreover, ACS5 and 2,3-oxidized squalene cyclase (OCS) were highly expressed in NZJ, while heme IX farnesyl transferase (IX-FIT) and ACS3 were significantly expressed in XZJ. The differential expression of ACS1, ACS2, HMGR, IX-FIT, SQS, and OCS was confirmed by real-time quantitative reverse transcription PCR. This study provides a new concept for the additional exploration of the molecular regulatory mechanism of terpenoid biosynthesis and data for the biotechnology of terpenoid production.
1. Introduction
Antrodia cinnamomea is a rare fungus that is endemic to Taiwan, China. It is a large edible and medicinal fungus that is a member of the Polyporaceae family [Citation1]. The fruiting body of A. cinnamomea rots on the inner wall of the decaying heartwood of Cinnamomum kanehirae, a key conservation tree species in Taiwan [Citation2]. Researchers found more than 70 compounds, including sesquiterpenes [Citation3], diterpenes [Citation4], triterpenes, and sterols, in the fruiting body and mycelia of A. cinnamomea [Citation5,Citation6]. Triterpenoids are the main active components of the fruiting body of A. cinnamomea, and they are the source of its bitterness [Citation7]. Currently, 34 ergostane triterpenoids and 10 lanosterol triterpenoids have been isolated from the fruiting body of A. cinnamomea. Ergostane (camphoric acid) compounds with more prominent biological activities are triterpenoids that are unique to A. cinnamomea. The content of triterpenoids in the fruiting bodies of wild A. cinnamomea > 10%, which is 1–3% higher than that of Ganoderma lucidum [Citation8]. Its medicinal uses include protecting the liver [Citation9]; reducing hypertension, hyperlipidemia, and hyperglycemia [Citation10]; combatting tumors [Citation11]; and serving as an antioxidant [Citation12].
The triterpene metabolism of A. cinnamomea is dominated by the mevalonate (MVA) pathway initiated by the two-molecule compound acetyl coenzyme A [Citation13]. Hydroxymethylglutaryl CoA synthase (HMGS), hydroxymethylglutaryl CoA reductase (HMGR), mevalonate pyrophosphate decarboxylase (MPD), squalene synthase (SQS), squalene epoxidase (SQE), and lanosterol synthase in the MVA pathway (LSS) and a series of ergostane synthetases are the key regulatory enzymes in different stages of the triterpene synthesis pathway of A. cinnamomea [Citation2].
Adjustment of the composition of culture medium can influence the yield of terpenoids in A. cinnamomea. The triterpene content of A. cinnamomea cultured in media that contained branch and leaf extracts of C. camphora was 30.78% higher than that of the control group [Citation14]. A high-pressure liquid chromatography chromatogram showed that the triterpene content of A. cinnamomea mycelia cultured in media amended with citrus peel increased significantly, and the triterpene content increased from 99.93 to 1,028.02 mg/L. This result showed that an extract of orange peel in liquid media promoted the production of triterpenoids in the A. cinnamomea mycelia [Citation15]. The addition of ginger extract to the solid media also significantly enhanced the yield of triterpenoids from A. cinnamomea and significantly increased its anticancer activity [Citation16]. In a study of the influence of birch bark on the yield of triterpenoids of the Chaga mushroom (Inonotus obliquus), the transcriptome of I. obliquus cultured in media with birch bark was sequenced using Illumina RNA-Seq. The expression of genes for 18 key enzymes in the terpene pathway had been influenced. The SQS (SQS2) involved in the terpene metabolic pathway was regulated at the transcriptional level after the addition of the birch bark and promoted the production of triterpenoids [Citation17]. However, regulation of the transcriptional level of triterpenoid biosynthesis in A. cinnamomea cultured in additives remains unknown.
In this study, homogenates of C. kanehirae and C. camphora shoots were used as additives to explore the effects of different culture conditions on the terpenoid metabolism of A. cinnamomea. The transcriptome was analyzed using Illumina HiSeq 2000 high-throughput sequencing, and the whole transcriptome was spliced and assembled. The differential expression was verified by quantitative real-time PCR (RT-qPCR) to reveal the genes related to the terpene metabolic pathway in the mycelia of A. cinnamomea.
2. Materials and methods
2.1. Fungal culture and growth conditions
The strain of A. cinnamomea (YAFAC008) that was used in this experiment was provided by the Yunnan Academy of Forestry & Grassland Science (Kunming, Yunnan, China). The strain of A. cinnamomea was inoculated in PDA, cultured in a constant temperature incubator at 26 °C and 60% humidity for 30 days, and then stored in a refrigerator at 4 °C.
2.2. Preparation of culture media
Cinnamomum kanehirae and C. camphora seedlings were planted in the greenhouse of Southwest Forestry University (Kunming, China) in 28 °C for two years. Clean twigs of C. kanehirae and C. camphora were collected for homogenization for addition to the culture media. The twig homogenates were obtained by a homogenizer and used as the additives. Solid media were PDA basal medium containing 1, 10, 20, and 40 g/L (W/V) twigs of C. kanehirae and C. camphora. The media were autoclaved and then cooled at room temperature. In addition, 0.5 cm2 of A. cinnamomea mycelium was evenly excavated from the edge of the mycelia and placed in a Petri dish with different substrates for dark culture at 26 °C for 14 days. The diameter of mycelial growth was measured every 2 days to observe the effects of different additives on mycelial growth.
2.3. Determination of the triterpene content in A. cinnamomea mycelia
Vanillin perchloric acid colorimetry was used to detect the effect of additives of C. kanehirae and C. camphora on the content of triterpenoids in A. cinnamomea. The method involved the addition of 40 mL of 75% ethanol to 1.0 g of dried A. cinnamomea mycelia, and 0.1 mL of the extract was removed and placed in a 7 mL test tube with a grinding plug, placed in boiling water for evaporation, and then 0.2 mL of a 5% solution of vanillin glacial acetic acid and 0.8 mL of perchloric acid were added. The mixture was subsequently placed in a constant temperature water bath at 70 °C for 15 min and then cooled to room temperature in ice water for 5 min. Next, 4 mL of ethyl acetate was added and mixed upside down. The absorbance at 550 nm was measured. Media that contained different additives without A. cinnamomea were also utilized as negative controls and subjected to the same method.
2.4. RNA extraction and sequencing library construction
Mycelial samples of A. cinnamomea cultured in the original medium (PDA) and media with the addition of C. camphora homogenate (XZJ) and C. kanehirae homogenate (NZJ) were collected for RNA extraction. RNA was extracted with a TransZol Up Plus RNA Kit (TransGen Biotech, Beijing, China), and approximately 3 μg of each sample was added to an NA 6000 Pico chip (Agilent Bioanalyzer; Agilent Technologies, Santa Clara, CA, USA) to identify the RNA. An NEB Next Ultra RNA Library Prep Kit for Illumina (NEB, Ipswich, MA, USA) was used to construct the sequencing library. The constructed library was sequenced by Personalbio Biotechnology Co., Ltd. (Shanghai, China) using an Illumina HiSeq 2000 sequencing platform (San Diego, CA, USA). The sequencing depth of the independent samples was 6 G.
2.5. Transcriptome assembly and functional annotation
The number and length of original data were counted after transcriptome sequencing. Clear reads were obtained from the original data by removing sequencing joints, repeated redundant sequences, and low-quality sequence data. The number, total length, Q20, n%, and GC% of the clear reads were then counted. Trinity software (Http//trinityrnaseq.github.io/) was used for de novo assembly. The reads with some length overlaps were connected into N-free fragment contigs, and then different contigs from the same transcript were connected to obtain non-redundant sequences that could not be extended at both ends. A sequence alignment method was used to quantify the sequence similarity of unigene, and non-redundant protein (NR), Gene Ontology (GO), and the Kyoto Encyclopedia of Genes and Genomes (KEGG) databases (E-value <10−5) were used for functional annotation of the A. cinnamomea unigenes.
2.6. Functional analysis of differential genes
The annotated transcripts were selected for analysis, and the expression of each transcript in different samples was calculated by fragments per kilobase of transcript per million mapped fragments (FPKM) to conduct a preliminary analysis of differential gene expression. DESeq was used for differential screening analysis, and the threshold was set as Q-value <0.001 and log2Ratio ≥1 (the specific algorithm and parameters had not been determined yet) to screen the differentially expressed genes (DEGs). GO enrichment analysis was conducted by goseq to screen out the significantly enriched GO pathway. With the KEGG pathway as the unit, a hypergeometric test was used to identify the pathway with the most significant enrichment in DEGs compared with the background of the assembled transcriptome.
2.7. Real time quantitative fluorescence PCR
Specific primers were designed to verify the results of transcriptome sequencing and the pattern of expression of the target gene (). The expression of the ACS1, ACS2, HMGR, heme IX farnesyl transferase (IX-FIT), SQS, and 2,3-oxidized squalene cyclase (OCS) genes were detected using an ABI 7300 Real Time PCR system (Applied Biosystems, Foster City, CA, USA). The PCR reaction system conditions were as follows: denaturation procedure (94 °C, 2 min), amplification and quantification procedure, which was repeated 40 times (94 °C, 15 s; 65 °C, 15 s; and 72 °C, 45 s), and finally, 72 °C for 10 min. The actin gene was used as an internal regulator of normal gene expression, and three independent biological replicates and three technical replicates per biological replicate were performed.
Table 1. Fluorescence quantitative detection of primer sequence.
3. Results
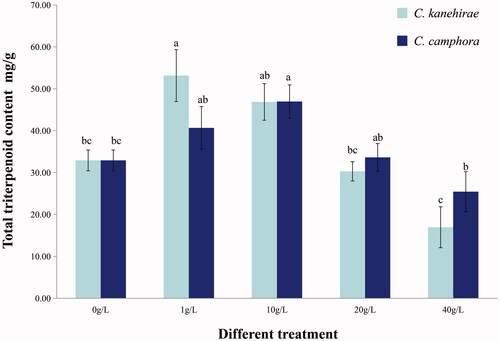
3.1. Effects of Cinnamomum twigs on the triterpene yield of Antrodia cinnamomea
The triterpenoid contents of the mycelia of A. cinnamomea were detected using vanillin perchloric acid colorimetry. This procedure showed that there were no triterpenoids in the media that contained different additives in the absence of A. cinnamomea. In the four different concentrations (1, 10, 20, and 40 g/L) compared with original PDA, the triterpene content of the mycelia of A. cinnamomea increased significantly in media with 1 g/L of C. kanehirae twig homogenate and decreased with higher concentrations of these homogenates. Similar results were obtained when the twig homogenates of C. camphora were added to the media. In addition, the highest triterpene content was detected in the mycelia of A. cinnamomea cultured in media with 10 g/L of twig homogenate of C. camphora ().
3.2. Assembly of the transcriptome data from Antrodia cinnamomea
The transcriptome of the mycelia of A. cinnamomea grown in three different types of media was sequenced using Illumina HiSeq 2000. Each treatment had three biological replicates, and three biological replicates were sequenced together, resulting in 146,777,202 bp raw data (raw reads). The original data were filtered to obtain 135,499,124 bp of effective data (clean reads) that comprised 92.3% of the clean reads to ensure the accuracy of subsequent biological information analyses. A total of 66,589 transcripts were obtained by de novo splicing and the assembly of valid data using the Trinity program, and the total length of the transcript was 212,645,633 bp, with an average length of 3193.4 bp and an N50 length of 4413 bp ().
Table 2. De novo assembly of transcriptome of Antrodia cinnamomea.
3.3. Functional annotation, classification, and metabolic pathway analysis of the unigenes from A. cinnamomea
The BLAST algorithm was used to compare and annotate the unigenes with the NR, GO, and KEGG databases to predict the biological functions of the coding genes of A. cinnamomea. The annotation information of the three databases was analyzed, and a total of 12,125 unigene annotations were successfully annotated simultaneously in all databases. The number of successful annotations of unigenes in the NR database was 6,061, which comprised 77.34% of the assembled transcripts. A total of 3,257 (41.56%) and 2,807 (35.82%) transcripts were annotated in the GO and KEGG databases, respectively, and 1,701 (21.78%) unigenes were annotated in all the databases.
NR is a taxonomic database used to obtain the similarity between the gene sequences of a species and its relatives and the functional information of the genes of a species. The 80,000 unigenes assembled were compared with the NR library through BLAST (evaluate <0.00001). A total of 6,061 unigenes were found to have similar sequences in the NR database ().
Figure 2. Non-redundant protein database functional classification of unigenes from Antrodia cinnamomea.
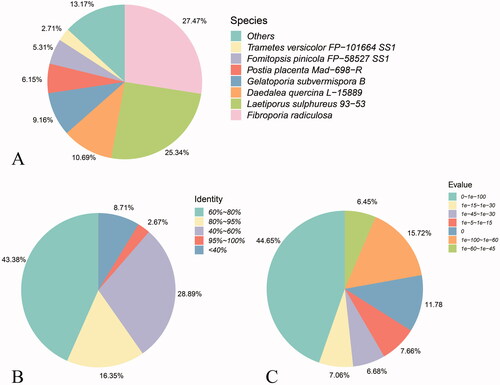
The results of matching annotation by species are shown in . The 6,061 unigenes of A. cinnamomea had varying degrees of homology with other known genes, and the five species with more annotation sequences were the brown rot fungi Fibroporia radiculosa, chicken of the woods (Laetiporus sulphureus), Daedalea quercina, Gelatoporia subvermispora, and Postia placenta, which comprised 27.47, 25.34, 10.69, 9.16, and 6.15%, respectively. The rest were distributed in more than 1,300 other species. As shown in the similarity distribution of matched sequences (), 43.38% of sequences demonstrated a similarity of 60–80%; 2.67% had a similarity of >80%, and 8.71% had a similarity of <60%. The E-value distribution () demonstrated that nearly half (44.65%) of the 6,061 unigenes annotated had E-values that were distributed in E−100–E−1, while 15.72% were distributed in E−100–E−60, comprising 11.78% when e = 0. The E-value and similarity distribution indicated that the matching degree of A. cinnamomea was higher in the NR library of NCBI.
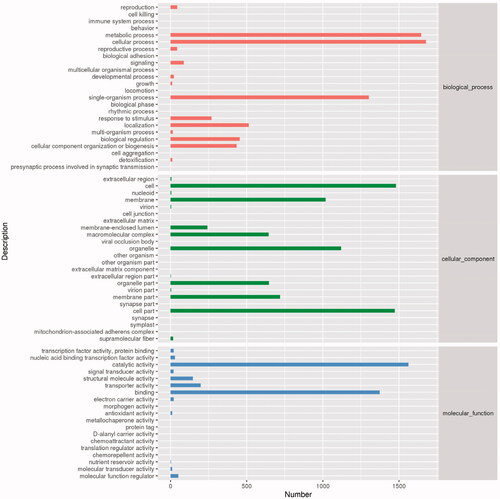
The GO function annotation was performed in accordance with the NR annotation. The GO gene classification included the annotation and functional analysis of a large number of annotated genes and their products in selected organisms. The biological significance of the genes could be understood by macroscopically analyzing the distribution characteristics of the gene function of A. cinnamomea. The analysis indicated that 3,257 unigenes were divided into three functional categories: cell component, molecular function, and biological process, and comprised 67 functional groups (). The results showed that cell components (35.82%) and biological processes (35.82%) were the most highly annotated, while cellular process, metabolic process, and single organization process were the most abundant in biological processes. Cell, cell part, and organelle composed the most of cell components, while cell killing, immune system process, cell junction, and extracellular matrix involved very few unigenes. Catalytic activity and binding were the most abundant in the molecular functional classification.
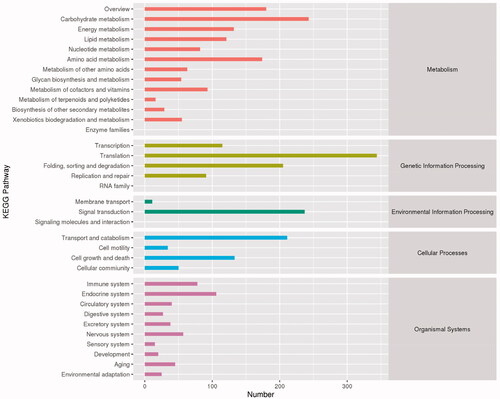
The unigenes in the KEGG database were compared, the metabolic pathway was analyzed in accordance with the annotation information, which enabled exploration of the metabolic pathway of gene products in cells and the function of gene products. The results showed that 2,870 unigenes (35.82%) were annotated. The statistical analysis of metabolic pathways that they may participate in or be involved with indicated that the unigenes of A. cinnamomea could be classified into five categories and 35 subclasses (). The results showed that among the five categories, metabolic-related pathways accounted for the most (37.14%), followed by organismal systems (28.57%), while environmental information processing (8.57%), cellular processes, and genetic information processing (8.57%) related pathways comprised 11.42 and 14.28%, respectively. The unigenes were further subdivided into subclasses. Metabolism-related pathways could be subdivided into 11 subclasses, which primarily included carbohydrate metabolism, amino acid metabolism, energy metabolism, and lipid metabolism. The other pathways related to environmental information processing, cellular process, and genetic information processing included three, four, and five subclasses, respectively, with most related to metabolic pathways, indicating that A. cinnamomea has strong metabolic activities during this period.
3.4. Differential expression analysis of terpene metabolism
In accordance with the KEGG terpene metabolic pathway and the data in , differential expression analysis was conducted on the database of A. cinnamomea. After adjustment, the transcripts that had p-values <0.05 and differential multiple (log2fc) > 1 were designated as the transcripts with significant differential expression. Among the three comparison groups with 359 differentially expressed transcripts, 1,102 DEGs were identified in the mycelia of A. cinnamomea cultured with C. camphora (PDA versus XZJ),with 128 upregulated and 231 downregulated. The number of transcripts in cells cultured with C. kanehirae (PDA versus NZJ) was 445, with 213 transcripts upregulated and 232 downregulated. In addition, 51 upregulated differential genes were found in the two groups (), indicating that adding different camphor tree culture media could regulate gene expression in the mycelia of A. cinnamomea and increase the number of DEGs under different culture conditions.
Figure 5. A Venn diagram of differentially expressed genes (DEGs) in the transcriptome of Antrodia cinnamomea. The sum of the numbers in each circle represents the total number of DEGs in the comparison combination, and the overlap of the circles represents the common DEGs between the two groups compared.
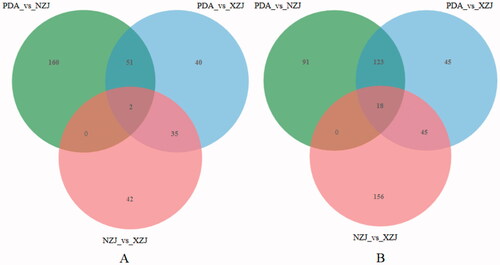
Table 3. Functional annotation of the unigenes in Antrodia cinnamomea.
Table 4. Genes of terpenoid metabolic biosynthesis in Antrodia cinnamomea.
3.5. Biosynthesis of terpenoids and related genes in the antrodia cinnamomea mycelia
A total of 23 transcriptional sequences of the mevalonate pathway involved in the synthesis of terpenoids were found in the transcriptome data of A. cinnamomea (). The number of DEGs in PDA versus NZJ was more than that in PDA versus XZJ, indicating that the addition of C. kanehirae homogenate has a substantial effect on the terpene metabolic pathway of A. cinnamomea. Genes involved in the non-mevalonate pathway were not identified, indicating a deletion in this pathway.
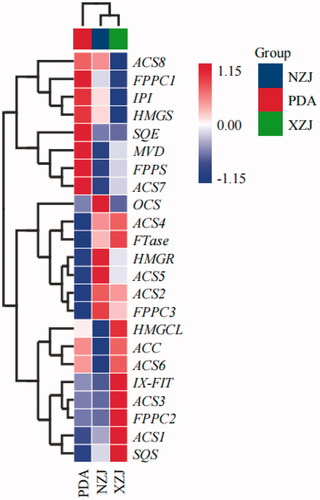
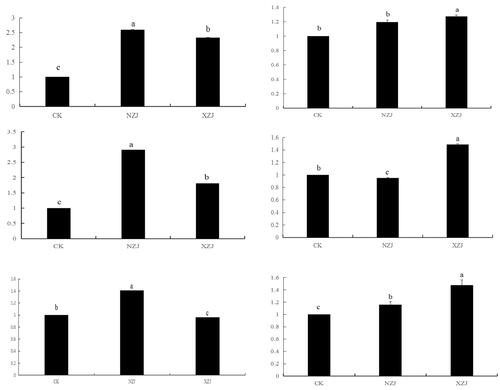
The expression profile data of terpene-related transcripts of A. cinnamomea were obtained to explore the expression of terpene-related transcripts under different growth conditions of A. cinnamomea. An expression interaction heatmap was drawn in accordance with the differential expression mode of the terpene metabolic pathway of A. cinnamomea cultured with a uniform slurry of young branches of C. camphora and C. kanehirae (). The expression profile data of the A. cinnamomea terpene transcripts demonstrated that the expression of these terpene transcripts cultured with C. camphora branch homogenate (XZJ) was significantly higher than culture of the fungus with C. kanehirae branch homogenate (NZJ). In the A. cinnamomea mycelia, the expression of upstream precursor genes ACS1 and ACS2 was significantly higher than that of the downstream specific genes FPPC3 (Sesquiterpene cyclase) and FTase. From the perspective of a precursor pathway, ACS2 and ACS4 were highly expressed in both treatments, indicating that they were involved in the transcriptional regulation of NZJ and XZJ. Moreover, HMGR, FTase, FPPC3, IX-FIT, and SQS were upregulated in the presence of NZJ and XZJ. Under XZJ culture conditions, the expression of FPPS was downregulated, whereas those of SQS and FPPC3 increased. Thus, the presence of camphor substrate positively regulated the biosynthesis of sesquiterpenes and triterpenes. In contrast, the expression of MPD was downregulated in the presence of terpene substrates, particularly in NZJ-cultured A. cinnamomea mycelia. MPD is a key enzyme in the mevalonate pathway. Thus, the addition of substrates was hypothesized to have promoted a negative feedback regulatory mechanism.
3.6. Validation of the RNA-Seq gene expression data by qRT-PCR
qRT-PCR was used to analyze the specific expression of terpene metabolism-related genes in A. cinnamomea cultured with C. camphora and C. kanehirae. The qRT-PCR analysis showed that the relative expression of six genes was consistent with the trend of transcriptome expression profile analysis (). In the presence of a terpene substrate, ACS1, ACS2, HMGR, SQS, and OCS were upregulated, indicating that they were involved in the positive regulation of terpene synthesis. In contrast, IX-FIT was downregulated in the presence of NZJ, whereas SQS and OCS were upregulated, suggesting that terpenoids, such as farnesyl diphosphate (FPP), exist in the twig residues of C. camphora. In conclusion, when terpene substrates exist in the mycelia of A. cinnamomea, several key response genes in the terpene pathway are regulated at the transcriptional level.
4. Discussion
The high-throughput sequencing platform Illumina RNA-Seq has been developed and used for transcriptome sequencing of model and non-model plants. Its reading sequence coverage is uniform, and the data quality is equal. It can be used to study DEGs among different species and detect new gene functions [Citation18]. The alteration of growth conditions can activate the production of fungal secondary metabolites, and it can also help to reveal the biosynthesis of bioactive compounds [Citation19]. The yield of triterpenoids could be increased in A. cinnamomea cocultured with Saccharomyces cerevisiae [Citation20]. Fermentation media with bark increased the accumulation of lanosterol in I. obliquus and Phellinus [Citation21,Citation22]. These results showed that the culture medium and method will influence the content of triterpenoids in fungi. However, the molecular mechanism remains unclear, and few studies have conducted transcriptome-level research on the influence of additives on fungal metabolism.
In this study, the mycelial transcriptome of A. cinnamomea cultured in original PDA and modified PDA with homogenates of C. camphora and C. kanehirae twigs was sequenced using an Illumina HiSeq 2000 platform. A total of 66,589 unigenes were obtained, and 23 genes related to the terpene pathway were identified. An analysis of the DEGs showed that the expression of HMGR in both NZJ and XZJ was higher than that in PDA, and the expression of HMGR in NZJ was significantly higher than that in XZJ. HMGR is a key rate-limiting enzyme in the MVA pathway. It catalyzes HMG-CoA to form mevalonate (MVA), accelerates the synthesis of downstream terpenes, and improves their yield [Citation23,Citation24]. The expression of HMGR in the A. cinnamomea mycelia differed significantly when carbon and nitrogen sources were added to different media and basic media, indicating that different culture conditions could induce the expression of HMGR [Citation25]. Zou et al. identified the involvement of HMGR (GenBank: AEX09818) in terpene synthesis in I. obliquus, and its expression was upregulated by I. obliquus in birch bark [Citation22]. This suggests that additives from the plant could stimulate the biosynthesis of triterpenes by upregulating the expression of HMGR.
Exploring the transcriptome-related genes of A. cinnamomea showed that it contained SQS, SQE, and OCS. The expression of SQS was upregulated in NZJ and XZJ, while that of SQE was significantly downregulated. Squalene is the common precursor of all triterpenoids. SQS catalyzes the synthesis of squalene from two molecules of farnesyl pyrophosphate. It is the first key enzyme in the synthesis pathway of triterpenoids. The activity and concentration of SQS determine the yield of triterpenoid products [Citation26,Citation27]. The upregulated expression of SQS in Panax ginseng significantly improved the biosynthesis of triterpenoids and sterols [Citation28]. The expression of SQS also increased in I. obliquus cultured with betulin, and the production of triterpenoids increased [Citation17].
Sesquiterpenes are catalyzed by the activity of sesquiterpene synthase on FPP, which plays an important role in the sesquiterpene skeleton structure [Citation29–31]. Lee et al. used transcriptomics to analyze the molecular mechanism of sesquiterpene synthesis in white rot fungi and identified that germacrene A synthase and trichodiene synthase were involved in sesquiterpene biosynthesis [Citation32]. Three sesquiterpene cyclase genes (FPPC1, FPPC2, and FPPC3) were identified based on the transcriptome of A. cinnamomea. FPPC2 and FPPC3 were 61% homologous, and the expression of FPPC2 and FPPC3 was upregulated after adding camphor as a substrate, while the expression of the precursor FPPS was downregulated in NZJ and XZJ.
In this study, the mycelia of A. cinnamomea cultured in medium with additives of C. kanehirae and C. camphora were used as materials to first determine the contents of triterpenes and perform a transcriptome analysis. We found that the additives stimulated the production of triterpenes in A. cinnamomea by regulating the key gene expression of HMGR and SQS involved in triterpene biosynthesis. This study will provide new information for the regulation of triterpene biosynthesis in A. cinnamomea. Future study will entail the separation of the compounds of the additives from C. kanehirae and C. camphora and confirmation that these chemicals stimulate triterpene production in A cinnamomea. We will also confirm the metabolic network of triterpene biosynthesis in A. cinnamomea.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Rao YK, Geethangili M, Tzeng YM. Development of a high-performance liquid chromatography method for the quantitative determination of bioactive triterpenoids in the extracts of Antrodia camphorata. Anal Methods. 2013;5(20):5724–5730.
- Lu MY, Fan WL, Wang WF, et al. Genomic and transcriptomic analyses of the medicinal fungus Antrodia cinnamomea for its metabolite biosynthesis and sexual development. Proc Natl Acad Sci USA. 2014;111(44):E4743–E4752.
- Yeh CT, Rao YK, Yao CJ, et al. Cytotoxic triterpenes from Antrodia camphorata and their mode of action in HT-29 human colon cancer cells. Cancer Lett. 2009;285(1):73–79.
- Toyomasu T. Recent advances regarding diterpene cyclase genes in higher plants and fungi. Biosci Biotechnol Biochem. 2008;72(5):1168–1175.
- Hsiao G, Shen MY, Lin KH, et al. Antioxidative and hepatoprotective effects of Antrodia camphorata extract. J Agric Food Chem. 2003;51(11):3302–3308.
- Joshi RA. Antrodia camphorata with potential anticancerous activities: a review. J Med Plant. 2017;5(1):284–291.
- Qiao X, Wang Q, Ji S, et al. Metabolites identification and multi-component pharmacokinetics of ergostane and lanostane triterpenoids in the anticancer mushroom Antrodia cinnamomea. J Pharm Biomed Anal. 2015;111:266–276.
- Geethangili M, Tzeng YM. Review of pharmacological effects of antrodia camphorata and its bioactive compounds. Evid Based Complement Alternat Med. 2011;2011(17):212641–21427X.
- Li ZW, Kuang Y, Tang SN, et al. Hepatoprotective activities of Antrodia camphorata and its triterpenoid compounds against CCl4-induced liver injury in mice. J Ethnopharmacol. 2017;206:31–39.
- Kuo YH, Lin CH, Shih CC. Antidiabetic and antihyperlipidemic properties of a triterpenoid compound, dehydroeburicoic acid, from Antrodia camphorata in vitro and in streptozotocin-induced mice. J Agric Food Chem. 2015;63(46):10140–10151.
- Du YC, Wu TY, Chang FR, et al. Chemical profiling of the cytotoxic triterpenoid-concentrating fraction and characterization of ergostane stereo-isomer ingredients from Antrodia camphorata. J Pharm Biomed Anal. 2012;58(1):182–192.
- Tien AJ, Chien CY, Chen YH, et al. Fruiting bodies of Antrodia cinnamomea and its active triterpenoid, antcin K, ameliorates N-nitrosodiethylamine-induced hepatic inflammation, fibrosis and carcinogenesis in rats. Am J Chin Med. 2017;45(1):1–26.
- Thapa HR, Naik MT, Shigeru O, et al. A squalene synthase-like enzyme initiates production of tetraterpenoid hydrocarbons in Botryococcus braunii race L. Nat Commun. 2016;7:11198.
- Lu ZM. Study on submerged culture of Antrodia cinnamomea its triterpenoids. Wuxi: Jiangnan University; 2009.
- Ma TW, Lai Y, Yang FC. Enhanced production of triterpenoid in submerged cultures of Antrodia cinnamomea with the addition of citrus peel extract. Bioprocess Biosyst Eng. 2014;37(11):2251–2261.
- Chen SY, Lee YR, Hsieh MC, et al. Enhancing the anticancer activity of Antrodia cinnamomea in hepatocellular carcinoma cells via cocultivation with ginger: the impact on cancer cell survival pathways. Front Pharmacol. 2018;9(780):780.
- Fradj N, Santos K, Montigny ND, et al. RNA-Seq de novo assembly and differential transcriptome analysis of chaga (Inonotus obliquus) cultured with different betulin sources and the regulation of genes involved in terpenoid biosynthesis. Int J Cell Sci Mol Biol. 2019;20(18):4334.
- Bastos DZ, Pimentel IC, de Jesus DA, et al. B.H. Biotransformation of betulinic and betulonic acids by fungi. Phytochemistry. 2007;68(6):834–839.
- Li G, Lou HX. Strategies to diversify natural products for drug discovery. Med Res Rev. 2018;38(4):1255–1294.
- Shu CH, Wu CJ, Hsiao WJ. Enhancement of triterpenoids production of Antrodia cinnamomea by co-culture with Saccharomyces cerevisiae. J Bioprocess Biotech. 2015;5(9):253.
- Zheng WF, Zhao YX, Zheng X, et al. Production of antioxidant and antitumor metabolites by submerged cultures of Inonotus obliquus cocultured with Phellinus punctatus. Appl Microbiol Biotechnol. 2011;89(1):157–167.
- Zou L, Sun T, Li D, et al. De novo transcriptome analysis of Inonotus baumii by RNA-seq. J Biosci Bioeng. 2016;121(4):380–384.
- Chen CC, Chyau CC, Hseu TH. Production of a cox-2 inhibitor, 2,4,5-trimethoxybenzaldehyde, with submerged cultured Antrodia camphorata. Lett Appl Microbiol. 2007;44(4):387–392.
- Shang CH, Zhu F, Li N, et al. Cloning and characterization of a gene encoding HMG-CoA reductase from Ganoderma lucidum and its functional identification in yeast . Biosci Biotechnol Biochem. 2008;72(5):1333–1339.
- Yuan XL, Xiao ZY, Chne LY, et al. Cloning and expression analysis of 3-hydroxy-3-methylglutaryl coenzyme a reductase gene in Antrodia camphorata (AcHMGR). Genomics Appl Biol. 2018;37(1):358–365.
- Haralampidis K, Trojanowska M, Osbourn AE. Biosynthesis of triterpenoid saponins in plants, in history and trends in bioprocessing and biotransformation. Adv Biochem Eng Biotechnol. 2002;75(2):31–49.
- Lee MH, Jeong JH, Seo JW, et al. Enhanced triterpene and phytosterol biosynthesis in Panax ginseng overexpressing squalene synthase gene. Plant Cell Physiol. 2004;45(8):976–984.
- Kim TD, Han JY, Huh GH, et al. Expression and functional characterization of three squalene synthase genes associated with saponin biosynthesis in Panax ginseng. Plant Cell Physiol. 2011;52(1):125–137.
- Agger S, Lopez-Gallego F, Schmidt-Dannert C. Diversity of sesquiterpene synthases in the basidiomycete Coprinus cinereus. Mol Microbiol. 2009;72(5):1181–1195.
- Wawrzyn GT, Bloch SE, Schmidt-Dannert C. Discovery and characterization of terpenoid biosynthetic pathways of fungi. Methods Enzymol. 2012;515(3):83–105.
- Ichinose H, Kitaoka T. Insight into metabolic diversity of the brown-rot basidiomycete Postia placenta responsible for sesquiterpene biosynthesis: semi-comprehensive screening of cytochrome P450 monooxygenase involved in protoilludene metabolism. Microb Biotechnol. 2018;11(5):952–965.
- Lee SY, Kim M, Kim SH, et al. Transcriptomic analysis of the white rot fungus Polyporus brumalis provides insight into sesquiterpene biosynthesis. Microbiol Res. 2016;182:141–149.