 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Many Apiospora species have been isolated from bamboo plants – to date, 34 bambusicolous Apiospora species have been recorded. They are known as saprophytes, endophytes, and plant pathogens. In this study, 242 bambusicolous Apiospora were isolated from various bamboo materials (branches, culms, leaves, roots, and shoots) and examined using DNA sequence similarity based on the internal transcribed spacer, 28S large subunit ribosomal RNA gene, translation elongation factor 1-alpha, and beta-tubulin regions. Nine Apiospora species (Ap. arundinis, Ap. camelliae-sinensis, Ap. hysterina, Ap. lageniformis sp. nov., Ap. paraphaeosperma, Ap. pseudohyphopodii sp. nov., Ap. rasikravindrae, Ap. saccharicola, and Ap. sargassi) were identified via molecular analysis. Moreover, the highest diversity of Apiospora was found in culms, and the most abundant species was Ap. arundinis. Among the nine Apiospora species, two (Ap. hysterina and Ap. paraphaeosperma) were unrecorded in Korea, and the other two species (Ap. lageniformis sp. nov. and Ap. pseudohyphopodii sp. nov.) were potentially novel species. Here, we describe the diversity of bambusicolous Apiospora species in bamboo organs, construct a multi-locus phylogenetic tree, and delineate morphological features of new bambusicolous Apiospora in Korea.
1. Introduction
Apiospora Sacc. (Apiosporaceae, Sordariomycetes, Ascomycota) was recognized and established with Ap. montagnei by Saccardo (1875), and 145 epithets of Apiospora have been listed in Index Fungorum (2022) [Citation1,Citation2]. Apiospora is a cosmopolitan fungus, reported from various sources such as plants, soil, air, and marine samples in tropical, subtropical, Mediterranean, temperate, and even cold regions [Citation3]. Moreover, they have been characterized as endophytes, saprobes, and plant pathogens (especially in Poaceae) [Citation4–7]. Morphologically, Apiospora is characterized by globose, subglobose to ellipsoid, oval, and obovoid conidia when observed in face view, lenticular in side view, and basauxic conidiogenous cells [Citation3]. The genus Apiospora has been observed to have Arthrinium-like morphs in the asexual state and is thus synonymized under Arthrinium species [Citation4,Citation8,Citation9]. However, differences in genetic, morphological, and ecological characteristics between the two genera were found by Pintos et al. [Citation3]; 76 species of Arthrinium have been synonymized under Apiospora, and the two genera have been completely separated [Citation3,Citation6,Citation10].
Bamboo plays a crucial role in global carbon cycling. It absorbs wastewater, and it is used in human economic activities, such as construction, furniture, food, and even medicine [Citation11]. Bamboo is also known as a good host, and more than 1300 bamboo ascomycetes (more than 120 families and 400 genera) have been described or recorded [Citation12]. Most bambusicolous fungi have been reported in bamboo organs, such as culms (665 species), leaves (216 species), sheaths (19 species), and branches (14 species), and the least number of fungi have been recorded in shoots, roots, and inflorescences [Citation12,Citation13]. According to previous research, the most commonly detected endophytic fungus in bamboo (Yushania brevipaniculata) is Arthrinium species (now including the genus Apiospora), comprising almost 50% of isolates, and it is also found in healthy bamboo leaves [Citation14]. Kim et al. [Citation15] isolated fungi (93 ascomycetes and 14 basidiomycetes) from bamboo chips with decayed parts and used them for the fungal decay test against bamboo [Citation15]. In the study, Ap. arundinis (=Ar. arundinis) was isolated as the second dominant species comprising 19.7% of the ascomycetes, and it contributed to the highest rate of weight loss (17.9%) against giant bamboo (Phyllostachys bambusoides) [Citation15]. However, a study of Apiospora diversity according to bamboo organs has not been conducted.
Approximately 70 bamboo species are distributed naturally or artificially in Korea, and the distribution area is estimated to occupy approximately 22,067 ha [Citation16]. However, studies on the diversity of bambusicolous fungi (including the bambusicolous Apiospora) in Korea are lacking. Currently, 17 Apiospora species have been reported in Korea. Among these, 14 Apiospora species were collected from marine environments (Ap. agari, Ap. arctoscopi, Ap. arundinis, Ap. fermenti, Ap. koreana, Ap. marii, Ap. marina, Ap. piptatheri, Ap. pusillisperma, Ap. rasikravindrae, Ap. sacchari, Ap. saccharicola, Ap. sargassi, and Ap. taeanensis). Three Apiospora species (Apiospora arundinis, Ap. camelliae-sinensis, and Ap. minutispora) have been collected from terrestrial environments, and only two Apiospora species have been reported in bamboo (Ap. arundinis and Ap. camelliae-sinensis) [Citation5,Citation6,Citation15,Citation17,Citation18].
This study aimed to investigate the bambusicolous Apiospora diversity in Korea with bamboo organ specificity and to report new Apiospora species (with unrecorded Apiospora) in Korea. To accurately identify the Apiospora species, four DNA molecular datasets of the internal transcribed spacer (ITS), 28S large subunit ribosomal RNA gene (LSU), translation elongation factor 1-alpha (TEF), and beta-tubulin (TUB) were used for phylogenetic analysis. Furthermore, a detailed analysis of cultural and microscopic characteristics was conducted.
2. Material and methods
2.1. Sampling and isolation
Bamboo materials (branches, culms, leaves, roots, and shoots) were collected from various bamboo forests in Korea (Figure 1S). A small piece of bamboo material was placed on a 2% malt extract agar (MEA) medium containing 0.01% streptomycin. Apiospora-like hyphae and spores were isolated continuously until they were pure isolates. The pure strains were stocked in glycerol 10% stock and stored at −20 °C in the Korea University Fungus Collection (KUC), Seoul, Korea. The strains examined in this study, including the type strains of novel Apiospora species candidates, were deposited at the National Institute of Biological Resources, Incheon, Korea (NIBR).
2.2. DNA extraction, polymerase chain reaction (PCR), and sequencing
Bambusicolous Apiospora strains were used for molecular identification. Genomic DNA was extracted from fungal mycelia using an AccuPrep Genomic DNA extraction kit (Bioneer, Daejeon, Korea) according to the manufacturer’s protocol. The AccuPower® PCR PreMix Kit (Bioneer) was used for PCR. PCR targeting ITS, LSU, TEF, and TUB regions. For the ITS region, ITS1F (or ITS5)/LR3 (or ITS4) primer sets were used [Citation19,Citation20]. For the LSU region, we used the LR0R/LR7 primer [Citation21]. To amplify the TEF region, 728F (or 983F)/1567R primer sets were used [Citation22,Citation23]. For TUB region, Bt2a (or T1)/Bt2b (or T2) primer sets were used [Citation24,Citation25]. All PCR products were checked by electrophoresis on a 1% agarose gel and purified using the AccuPrep DNA Purification Kit (Bioneer). DNA sequencing was conducted by Cosmo Genetech (Seoul, Korea). All new sequences have been deposited in GenBank.
2.3. Phylogenetic analysis
All obtained sequences were assembled, proofread, and edited using Geneious Prime 2022.1.1 (Biomatter, Ltd., Auckland, New Zealand). The edited sequences were aligned with reference sequences of Apiospora, Arthrinium, and related genera downloaded from the GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) using MAFFT 7.450 [Citation26,Citation27]. The ambiguous alignments were manually adjusted, and maximum likelihood (ML) analysis was performed using RAxML v. 8 with the GTR + G model with 1000 bootstrap replicates [Citation28]. MrBayes (MB) analysis was carried out using MrBayes v. 3.2.6, with the best model selected for each ITS, LSU, TEF, and TUB dataset using jModeltest v. 2.1.10 [Citation29,Citation30]. To achieve stationary equilibrium, five million trees were generated, and the trees were sampled every 1000th generation. Posterior probabilities (PP) were calculated in the majority rule consensus tree after discarding the first 25% of the trees as burn-in. All analyses were performed using Geneious Prime software 2022.1.1 (https://www.geneious.com/prime/).
2.4. Morphological observation
The culture characteristics and growth rates of Apiospora were observed on the potato dextrose agar (PDA, Difco, Detroit, USA), MEA, and oatmeal agar (OA, Difco) media at 15 °C, 20 °C, and 25 °C in darkness for 2 weeks. The colony form, elevation, margin, presence of aerial mycelia, the color of mycelia and medium, and sporulation were recorded. Color-corresponding codes were determined according to the Munsell color chart (Munsell Color, 2009). Growth rates were measured every 24 h, and each measurement was performed in triplicates. Microscopic characteristics were observed on water agar medium (WA, Bacto agar (Difco) 15 g, distilled water 1000 mL) using an Olympus BX51 light microscope (Olympus, Tokyo, Japan) with a DP20 microscope camera (Olympus). The shape, size, and color of the conidiophores, conidiogenous cells, conidia, and hyphae were observed and recorded. Ultra-high-resolution scanning electron microscopy (UHR SEM, Hitachi SU-70, Hitachi, Tokyo, Japan) was used to observe the detailed morphological characteristics.
3. Results
3.1. Diversity of bambusicolous Apiospora in Korea
A total of 108 bamboo samples were collected from 20 bamboo forests in Korea (Figure 1S). The collected bamboo materials were composed of 33 branches, 44 culms, 14 leaves, 13 roots, and four shoots, and were used as fungal isolation sources. As a result, 242 bambusicolous Apiospora strains were isolated and identified based on the DNA sequence similarity of ITS, LSU, TEF, and TUB regions against the NCBI database (http://www.ncbi.nlm.nih.gov/blast). Based on sequence similarity, the Apiospora strains were identified as nine Apiospora species (Ap. arundinis (181 strains), Ap. camelliae-sinensis (17 strains), Ap. hysterina (two strains), Ap. rasikravindrae (31 strains), Ap. saccharicola (two strains), Ap. sargassi (one strain), Ap. paraphaeosperma (two strains), Ap. lageniformis sp. nov. (four strains), and Ap. pseudohyphopodii sp. nov. (two strains)). Figure 2S shows that the diversity of Apiospora was the highest in the culm, followed by the branch, and the most abundant species was Ap. arundinis, which accounted for >74% of the total isolates, followed by Ap. rasikravindrae (13%) and Ap. camelliae-sinensis (7%), respectively. The portion of Ap. camelliae-sinensis and Ap. rasikravindrae was higher in the bamboo branch but lower in the culm (Figure 2S). A few Apiospora species have been isolated from leaves, roots, and shoots. Apiospora arundinis was isolated from the highest proportion of bamboo tissues. Apiospora sargassi has only been isolated from the shoot tissues.
Apiospora hysterina strains were isolated from bamboo branches and Ap. paraphaeosperma strains were isolated from culms. The strains of Ap. pseudohyphopodii sp. nov. was isolated from bamboo culms, and Ap. lageniformis sp. nov. was isolated from the branches and culms. According to the present study, two species (Ap. hysterina and Ap. paraphaeosperma) and two novel species (Ap. lageniformis sp. nov. and Ap. pseudohyphopodii sp. nov.) have been recognized as new candidate species in Korea. Thus, phylogenetic and morphological analyses were performed for accurate taxonomic evaluation.
3.2. Phylogenetic analysis
The multigene alignments (ITS, LSU, TEF, and TUB combined datasets) contained 151 reference strains, and 10 new isolated strains in this study with 3717 characters, including gaps, were analyzed using ML and MB methods. The multigene alignments (ITS, LSU, TEF, and TUB combined datasets) contained 151 reference strains, and 10 new isolated strains in this study with 3717 characters, including gaps, were analyzed using ML and MB methods (). In MB analysis, ITS and LSU sequence alignments were assigned as GTR + I+G to the best-fit model, and TEF and TUB were assigned as GTR + G and HKY + I+G, respectively. Both ML and MB trees showed similar tree topologies, and the ML tree is represented. Two new Apiospora species (Ap. lageniformis sp. nov. and Ap. pseudohyphopodii sp. nov.) were distinct from other Apiospora clades and were clustered as monophyletic groups, respectively with high support (1/100, PP/bootstrap value (BS)) (). Although Ap. hysterina KUC21437 and KUC21438 formed a monophyletic group with Ap. hysterina ICPM 6889 and Ap. hysterina AP29717, they were not distinguished from Apiospora sasae CPC 38165 and Ap. yunnana MFLUCC 18-1102. Furthermore, Ap. paraphaeosperma KUC21488 and KUC21688 were grouped together with Ap. paraphaeosperma GUCC 10126 and MFLUCC 13-06044, but the resolution was low in the concatenated tree (). A morphoanatomical analysis is needed to interpret the low resolution of the two unrecorded Apiospora species.
Figure 1. ML tree based on ITS, LSU, TEF, and TUB concatenated datasets. The node numbers indicate the Bayesian posterior probabilities (PP) > 0.70 and ML bootstrap support (BS) > 70% as PP/BS. The novel Apiospora cultures examined in this study are shown in bold-face orange color. The unrecorded species are denoted by a green color. Type materials indicated by “T”.
Table 1. Strain informations included in the phylogenetic analyses.
3.3. Taxonomy
Apiospora lageniformis S.L. Kwon & J.J. Kim, sp. nov. ()
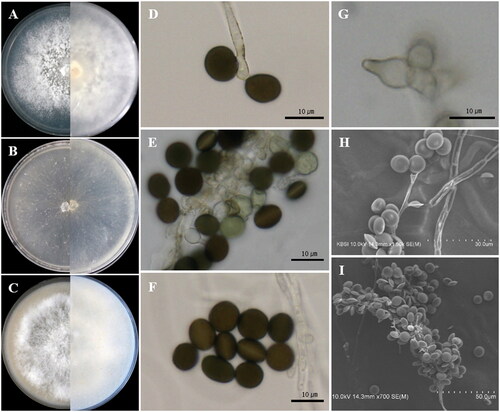
Figure 2. Apiospora lageniformis (KUC21686). (A) PDA; (B) MEA; (C) OA; (D, E) conidiogenous cell with conidia; (F) conidia; (G) lageniform conidiogenous cell; (H, I.) clustered conidia under UHR-SEM.
MycoBank: MB845439
Type: KOREA, Jeollabuk-do, Damyang-gun, 32°34′27.4′′N, 124°52′17.8′′E, isolated from the culm of Phyllostachys nigra var. henonis, Apr. 2021, S.L. Kwon (NIBRFGC000509393 = KUC21686).
Etymology: “lageniformis” refer to the lageniform shape of the conidiogenous cell.
Culture characteristics: PDA, colonies irregular form, flat, mycelium moderate, concentrically spreading, margin filiform; mycelia white; sporulation observed after 7 days at 15 °C on hyphae; pigment not observed. MEA, colonies circular form, flat, mycelium low, concentrically spreading with sparse aerial mycelium, margin entire; mycelia hyaline to white colored; sporulation observed after 7 days at all temperatures on hyphae; pigment absent. OA, colonies circular form, mycelium abundant, fluffy, downy, crateriform, thick, concentrically spreading with abundant aerial mycelium, margin entire; mycelia white; sporulation not observed; pigment absent.
Colony diameters – 15 °C PDA 6.4–7 cm/14 days, MEA 6.5–6.6 cm/14 days, OA 5.1–5.5 cm/14 days; 20 °C PDA 7 cm/13 days, MEA 7 cm/12 days, OA 7 cm/13–14 days; 25 °C PDA 7 cm/13 days, MEA 7 cm/9 days, OA 7 cm/12–13 days.
Asexual morphology: Conidiophores are reduced to conidiogenous cells. Conidiogenous cells aggregated in a cluster on hyphae, basauxic, polyblastic, hyaline, lageniform, 8.0–10.5(–12) × 4.0–5.0 µm, apical neck 3.5–5.5 µm long, basal part 2.8–7.2 µm long. Conidia green to dark brown, surface smooth, globose to ellipsoid in surface view, (7.8–)8.1–9.0(–9.5) × (6.8–)7.5–8.5(–9.0) µm ( = 8.6 × 8.0 µm, n = 30); lenticular in side view, with equatorial slit, (7.0–)8.0–9.5(–9.5) × (5.3–)6.0–7.0(–7.5) µm (
= 8.6 × 6.4 µm, n = 30). Mycelium smooth, hyaline, branched, septate, 2.0–4.0 µm diam.
Additional materials examined: KOREA, Jeollabuk-do, Damyang-gun, 32°34′27.4′′N, 124°52′17.8′′E, isolated from the culm of Phyllostachys nigra var. henonis, Apr. 2021, S.L. Kwon (NIBRFGC000509394 = KUC21687); KOREA, Jeollabuk-do, Gochang-gun, 35°25′50.9′′N, 126°42′16.9′′E, isolated from a branch of Phyllostachys pubescens, Mar. 2021, S.L. Kwon (NIBRFGC000509391 = KUC21681 and NIBRFGC000509392 = KUC21685).
Remarks: The Ap. lageniformis sp. nov. is characterized by a lageniform conidiogenous cell. This species is closely related to Apiospora jiangxiensis LC4577 (M. Wang & L. Cai) Pintos & P. Alvarado (over 100% similarity in the ITS region, 100% in the LSU region, 99.77% in the TEF region, and 97.92% in the TUB region). However, they can be distinguished by phylogenetic analysis with high bootstrap values (1/100, PP/BS). In the original description, Ap. jiangxiensis LC4577 had luteous to sienna pigments on colonies and media [Citation7]. However, no pigments were observed in the Ap. lageniformis sp. nov. Furthermore, the growth rate of Ap. jiangxiensis LC4577 (9 cm/10 days, at 25 °C on PDA) was faster than Ap. lageniformis sp. nov. KUC21686 at 25 °C on PDA (7 cm/13 days) [Citation7]. Apiospora lageniformis sp. nov. also is closely related to Apiospora obovata (M. Wang & L. Cai) Pintos & P. Alvarado, and Ap. arctoscopi (S.L. Kwon, S. Jang & J.J. Kim) S.L. Kwon & J.J. Kim in concatenate phylogeny (). However, Ap. obovata has obovoid, elongated to ellipsoidal conidia (size 16–31 × 9–16 µm), and A. arctoscopi has globose to elongated ellipsoid (in surface view, 9.5–13 × 7.5–12 µm) conidia (in lenticular side view, 5.5–7.5 µm) [Citation5,Citation7], which are different conidia characteristics of Ap. lageniformis. Apiospora arctoscopi also has different conidiogenous cell shapes (cylindrical, sometimes ampulliform) [Citation5].
Apiospora pseudohyphopodii S.L. Kwon & J.J. Kim, sp. nov. ()
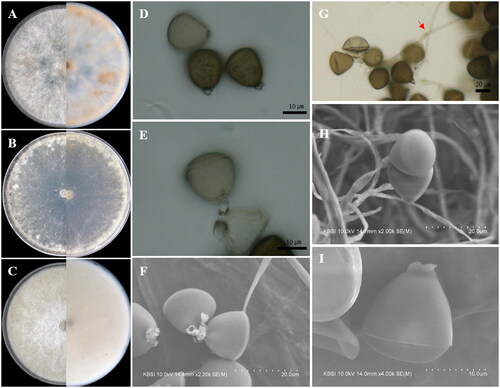
Figure 3. Apiospora pseudohyphopodii (KUC21680). (A) PDA; (B) MEA; (C) OA; (D, E) conidiogenous cell with conidia; (F) conidia generated on WA medium under light microscope; (G) lobed hyphopodia; (H) conidiogenous cell with ellipsoidal conidia under UHR-SEM; (I) lobed hyphopodia under UHR-SEM.

MycoBank: MB845440
Type: KOREA, Jeollabuk-do, Gochang-gun, 35°25′50.9′′N, 126°42′16.9′′E, isolated from a branch of Phyllostachys pubescens, Mar. 2021, S.L. Kwon (NIBRFGC000509202 = KUC21680)
Etymology: Named after its morphological similarity to Apiospora hyphopodii.
Culture characteristics: PDA, colonies circular form, flat, mycelium dense around the center and become sparse at the margin, concentrically spreading with abundant aerial mycelium, margin filiform; mycelia white around the center, fading to hyaline at the margin; sporulation observed after 7 days at 15 °C on hyphae; yellow (2.5Y, 7/8) pigment diffused after 5 days, and becoming converted to dark olive gray (5Y, 3/2) pigment from the center in reverse. MEA, colonies circular form, flat, mycelium low, concentrically spreading with sparse aerial mycelium, margin filiform; mycelia white colored; sporulation observed around plug after 7–8 days at 15 °C; pigment absent. OA, colonies circular form, flat, mycelium abundant, dense, concentrically spreading with sparse aerial mycelium, margin entire; mycelia white; sporulation not observed; pigment absent.
Colony diameters – 15 °C PDA 3.2–3.5 cm/14 days, MEA 1.9–2.2 cm/14 days, OA 7 cm/12–13 days; 20 °C PDA 5.2–6.2 cm/14 days, MEA 4–4.3 cm/14 days, OA 7 cm/5–6 days; 25 °C PDA 7 cm/9 days, MEA 7 cm/11–12 days, OA 7 cm/5 days.
Asexual morphology: Conidiophores are reduced to conidiogenous cells. Conidiogenous cells solitary on hyphae, hyaline, cylindrical, 9.5–13(–24) × 4.5–5.5 µm. Conidia were brown, smooth, globose to ellipsoid, sometimes polygonal or irregular, 20–25(–26) × 18–23 µm ( = 22.4 × 21.1 µm, n = 37). Elongated conidia brown, smooth, obovoid, clavate, (25–)27–40(–44) × 12–20(–22) µm in size. Hyphopodia blackish, lobed, irregular in shape, resembling coral and sea squirt, 20–35(–42) × 5–35 µm. Mycelium smooth, hyaline, branched, and septate.
Additional material examined: KOREA, Jeollabuk-do, Gochang-gun, 35°25′50.9′′N, 126°42′16.9′′E, isolated from a branch of Phyllostachys pubescens, Mar. 2021, S.L. Kwon (NIBRFGC000509389 = KUC21684).
Remarks: The Apiospora pseudohyphopodii sp. nov. is closely related to Apiospora pseudoparenchymatica LC7234 (over 96.2% similarity in the ITS region, 99.52% in the LSU region, 92.92% in the TEF region, and 93.62% in the TUB region) and Ap. hyphopodii MFLUCC 15-003 (over 98.68% similarity in the ITS region) in the phylogenetic analysis (). This species is characterized by blackish-lobed hyphopodia and large and elongated conidia. Hyphopodia have also been observed in Ap. hyphopodii MFLU 15-0383 [31]. However, Ap. pseudohyphopodii sp. nov. KUC21680 has larger conidia (20–25(–26) × 18–22.5 µm (x = 22.5 × 21.2 µm, n = 37)) than Ap. hyphopodii MFLU 15-0383 (5–10 × 4–8 µm (x = 6.5 × 5.6 µm, n = 20)) [Citation31]. The conidia of Ap. pseudoparenchymatica are similar in size to those of Ap. pseudohyphopodii sp. nov. KUC21680. However, they were clearly distinguished based on their phylogenies. Also, the growth rate of Ap. pseudohyphopodii sp. nov. KUC21680 (7 cm/9 days at 25 °C on PDA) is slower than Ap. pseudoparenchymatica (9 cm/8 days at 25 °C on PDA) [Citation7].
Apiospora hysterina (Sacc.) Pintos & P. Alvarado, Fungal Systematics and Evolution 7:206 (2021) [MB837743] ().
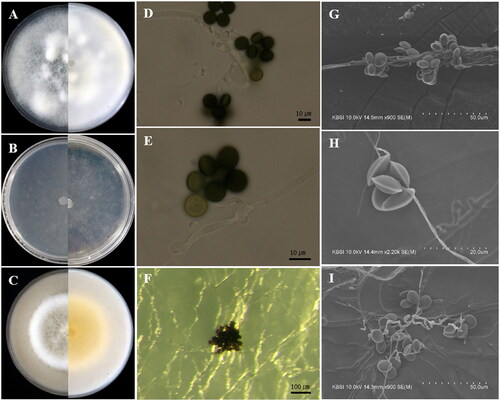
Figure 4. Apiospora hysterina (KUC21437). (A) PDA; (B) MEA; (C) OA; (D) conidia; (E, G) conidiogenous cell with conidia; (F, H, I) conidia under UHR-SEM.
Culture characteristics: PDA, colonies circular form, flat, mycelium moderate, concentrically spreading with abundant aerial mycelium, margin entire; mycelia white; sporulation observed after 7–10 days at 15 °C and 20 °C on hyphae; reddish yellow (5YR, 7/8) pigment partially observed after 11 days. MEA, colonies circular form, flat, mycelium low, concentrically spreading with aerial mycelium, margin entire; mycelia hyaline to white colored; sporulation observed after 7–10 days at all temperatures on hyphae; pigment absent. OA, colonies circular form, flat, mycelium concentrically spreading with abundant aerial mycelium, margin entire; mycelia white; sporulation observed after 7–10 days at 20–25 °C on hyphae; pigment absent.
Colony diameters – 15 °C PDA 5.4–5.8 cm/14 days, MEA 4.8–4.9 cm/14 days, OA 5.5–6.8 cm/14 days; 20 °C PDA 7 cm/9–10 days, MEA 7 cm/11–12 days, OA 7 cm/9–10 days; 25 °C PDA 7 cm/7 days, MEA 7 cm/8 days, OA 7 cm/7 days.
Asexual morphology: Conidiophores basauxic, polyblastic, hyaline to pale brown, septate or not, smooth or finely roughened with granular pigments, cylindrical, straight or flexuous, 10–25 × 2–3.5 µm, sometimes exceeding 98 µm long. Conidia brown to dark brown, surface smooth, finely roughened, globose to subglobose in surface view, 15.0–18.0 × (13.2–)14.0–16.5(–17.5) µm ( = 16.3 × 15.7 µm, n = 30); obovoid with a horizontal scar at the edge in side view, 15.0–18.0 × (11.5–)13.0–16(–17.5) µm (
= 16.7 × 14.9 µm, n = 50).
Specimen examined: KOREA, Chungcheongnam-do, Taean-gun, 36°29′51.0′′N, 126°21′41.5′′E, isolated from the branch of Phyllostachys bambusoides, Feb. 2020, S.L. Kwon (NIBRFGC000506558 = KUC21437 and NIBRFGC000509388 = KUC21438).
Remarks: The microscopic morphologies of Ap. hysterina KUC21437 and KUC21438 are well-matched with the original description. The former has longer conidiophores exceeding 98 µm and obovoid conidia with a horizontal scar resembling Ap. hysterina ICMP 6889 [Citation32]. The diffused pigment of Ap. hysterina ICMP 6889 was observed on MEA [Citation32]. However, the pigment of Ap. hysterina KUC21437 was not observed on the MEA medium but was observed on the PDA medium. The obovoid shape of conidia of Ap. hysterina are similar to those of Apiospora yunnana (D. Q. Dai & K.D. Hyde) Pintos & P. Alvarado, and Ap. sasae Crous & R.K. Schumach, and they are closely related in the concatenated phylogenetic tree (). However, the long conidiophores and small conidia of Ap. hysterina KUC21437 differs from Ap. yunnana [Citation33]. In the case of Apiospora sasae, it is morphologically similar to Ap. hysterina by producing subglobose, polygonal to urceolate (uniform) conidia ((16–)17–18(–20) × (15–)16–17(–19) µm) [Citation34]. However, this species can be distinguished by the septate and long conidiophores of Ap. hysterina KUC21437.
Apiospora paraphaeosperma (Senan. & K.D. Hyde) Pintos & P. Alvarado, Fungal Systematics and Evolution 7:206 (2021) [MB837705] ()
Figure 5. Apiospora paraphaeosperma (KUC21488). (A) PDA; (B) MEA; (C) OA; (D, E) conidiogenous cell with conidia; (F) conidia generated on WA medium under light microscope; (G–I) conidiogenous cell with conidia under UHR-SEM.
Culture characteristics: PDA, colonies circular form, mycelium thick, fluffy, concentrically spreading, margin entire; mycelia white, partially yellow; sporulation not observed; pigment absent. MEA, colonies circular form, flat, mycelium low, margin entire; mycelia hyaline to white colored; sporulation observed after 8–10 days at 20–25 °C on hyphae; pigment absent. OA, colonies circular form, flat, mycelium thick, fluffy, concentrically spreading with abundant aerial mycelium, margin entire; mycelia white, partially yellow; sporulation not observed; Yellow (2.5Y, 8/8) pigment partially diffused in media.
Colony diameters – 15 °C PDA 5.2–5.3 cm/14 days, MEA 4.3–4.5 cm/14 days, OA 4.0–4.2 cm/14 days; 20 °C PDA 7.0 cm/13 days, MEA 5.3–5.8 cm/14 days, OA 5.5–6.0 cm/14 days; 25 °C PDA 7.0 cm/11–12 days, MEA 7.0 cm/12–13 days, OA 6.5-7.0 cm/14 days.
Asexual morphology: Conidiophores are reduced to conidiogenous cells. Conidiogenous cells aggregated in clusters on hyphae, basauxic, polyblastic, hyaline, cylindrical, and ampulliform, 3.0–5.1(–8.7) × 1.5–3.0 µm, elongated conidiogenous cells length (11–)15–25(–34) µm. Conidia green to brown, surface smooth, globose to subglobose, 9.5–12.0 × 8.0–11.0 µm ( = 10.9 × 9.8 µm, n = 47) in surface view; lenticular in side view, with equatorial slit, 7.5–9.0 µm wide (
= 8.12 µm, n = 37) in side view, a slightly elongated cell was observed. Mycelium smooth, hyaline, branched, septate, 1.5–2.5 µm diam.
Specimen examined: KOREA, Jeju-do, Seogwipo-si, 33°15′26.4′′N, 126°21′11.2′′E, isolated from a culm of bamboo, 2018, J.J. Kim, (NIBRFGC000509203 = KUC21488 and NIBRFGC000509390 = KUC21688).
Remarks: In the original description, Ap. paraphaeosperma MFLUCC 13-0644 had a long conidiogenous cell (25–30 × 4–6 µm) [Citation35]. Although the conidiogenous cells of Ap. paraphaeosperma KUC21488 usually were observed at an average of 3.0–5.1(–8.7) µm long, sometimes the elongated conidiogenous cells are also observed ((11–)15–25(–34) µm long). This species is closely related to Apiospora rasikravindrae (Shiv M. Singh et al.) Pintos & P. Alvarado, and Apiospora marina (S.L. Kwon, S. Jang & J.J. Kim) S.L. Kwon & J.J. Kim in the concatenated phylogenetic analysis. However, they could be distinguished by the presence or absence of elongated conidiogenous cells in Ap. paraphaeosperma.
4. Discussion
In this study, 242 bambusicolous Apiospora strains were isolated from various bamboo organs and identified based on their DNA similarity against the NCBI database. As a result, in the bamboo organs, the highest Apiospora diversity was detected on the culms (seven species), followed by branches (six species), leaves (two species), shoots epidermis (two species), and roots (one species) (Figure 2S). The finding that the most diverse Apiospora were found in bamboo culms is consistent with the previously reported result that most bambusicolous Apiospora species have been isolated from bamboo culms (23 species/34 species of total bambusicolous Apiospora) () [Citation4,Citation10,Citation31–33,Citation35–45].
Table 2. List of bambusicolous Apiospora in worldwide.
So far, only Ap. rasikravindrae species have been reported in bamboo shoots by Majeedano et al. [Citation46]. In addition, no studies have reported on the isolation of Apiospora species from bamboo roots (). However, in this study, Ap. arundinis was isolated from all organs, including shoots and roots. In addition, this species had the highest abundance (74% of the total isolates) among the bambusicolous Apiospora species (Table 1S).
New records were identified based on morphological and phylogenetic analyses. The DNA barcode set (ITS, LSU, TEF, and TUB regions) was used in the phylogenetic analysis to distinguish them from cryptic species. In the case of Ap. pseudohyphopodii sp. nov., it is difficult to distinguish between them using only morphology. However, they were clearly distinguished in the phylogenetic analysis, with high bootstrap values (). The Ap. pseudohyphopodii sp. nov. is morphologically noted to have hyphopodia and large conidia (). Hyphopodia structures were also observed in the species Ap. hyphopodii within the genus Apiospora [Citation31]. However, Ap. hyphopodii could be distinguished by having smaller conidia than Ap. pseudohyphopodii sp. nov. The conidia size of Ap. pseudohyphopodii sp. nov. (globose to ellipsoid, sometimes polygonal or irregular, 20–25(–26) × 18–22.5 µm (x = 22.5 × 21.2 µm, n = 37)) is similar to Ap. neogarethjonesii (globose to subglobose, 20–35 × 15–30 µm), Ap. pseudoparenchymatica (globose to subglobose, 13.5–27 × 12–23.5 µm), and Ap. yunnana (globose to obovoid, 17.5–26.5 × 15.5–25 µm) [Citation7,Citation33,Citation42]. However, they could be distinguished by the shape of the conidia, the presence or absence of hyphopodia, and phylogeny. The Ap. lageniformis sp. nov. is closely related to Ap. jiangxiense (M. Wang & L. Cai) Pintos & P. Alvarado, Ap. obvata (M. Wang & L. Cai) Pintos & P. Alvarado, and Ap. arctoscopi (S.L. Kwon, S. Jang & J.J. Kim) S.L. Kwon & J.J. Kim in concatenate phylogeny (). However, they could be distinguished by culture characteristics, growth rates, conidia size, and conidiogenous cell shape. The Ap. lageniformis sp. nov. is characterized by basauxic, polyblastic, and lageniform conidiogenous cells. The other two unrecorded species, Ap. hysterina and Ap. paraphaeosperma, could also be distinguished from cryptic species and identified as a new record species in this study, but both morphological and phylogenetic analyses are needed.
To date, 34 Apiospora species have been reported in bamboo materials worldwide (). In contrast, only two bambusicolous Apiospora species have been reported in Korea (Ap. arundinis and Ap. camelliae-sinensis) [Citation15,Citation17]. In the present study, nine Apiospora species contained two unrecorded species (Ap. hysterina and Ap. paraphaeosperma), five recorded species (Ap. arundinis, Ap. camelliae-sinensis, Ap. rasikravindrae, Ap. sargassi, and Ap. saccharicola), and two novel species (Ap. pseudohyphopodii sp. nov. and Ap. lageniformis sp. nov.) were found in bamboo forests. Two previously unrecorded species have been reported from bamboo materials in New Zealand (Ap. hysterina), Spain (Ap. hysterina), and Thailand (Ap. paraphaeosperma) [Citation32,Citation35]. Moreover, one recorded species, Ap. rasikravindrae has been reported in bamboo in China [Citation7]. However, the other two recorded species (Ap. sargassi and Ap. saccharicola) have not been reported in bamboo until now; thus, this is the first report of these species isolated from bamboo materials.
Research on bambusicolous fungi may provide opportunities to control bamboo pathogens and promote bamboo cultivation [Citation47]. However, the ecological roles of most of the Apiospora remain unknown. Therefore, Apiospora diversity and their ecological roles need to be explored further. This study will serve as a basis for the taxonomic study of Apiospora and is expected to be the groundwork for potentially determining the diversity of Apiospora in the bamboo forests of Korea.
Supplemental Material
Download MS Word (220.2 KB)Acknowledgment
The authors are grateful to Dr. Songjin Lee (Bamboo Resource Research Institute, Damyang-gun, Korea) for help in collecting and identifying the bamboo materials.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Correction Statement
This article was originally published with errors, which have now been corrected in the online version. Please see Correction (http://dx.doi.org/10.1080/12298093.2023.2239652)
Additional information
Funding
References
- Saccardo P. Conspectus generum pyrenomycetum italicorum additis speciebus fungorum venetorum novis vel criticis, systemate carpologico dispositorum. Atti della società Veneziana-Trentina-Istriana di. Scienze Naturali. 1875;4:77–100.
- Index Fungorum. 2022. http://www.indexfungorum.org/Names/Names.asp
- Pintos Á, Alvarado P. Phylogenetic delimitation of Apiospora and Arthrinium. Fungal Syst Evol. 2021;7:197–221.
- Crous PW, Groenewald JZ. A phylogenetic re-evaluation of Arthrinium. IMA Fungus. 2013;4(1):133–154.
- Kwon SL, Park MS, Jang S, et al. The genus Arthrinium (Ascomycota, Sordariomycetes, Apiosporaceae) from marine habitats from Korea, with eight new species. IMA Fungus. 2021;12(1):1–26.
- Kwon SL, Cho M, Lee YM, et al. Two unrecorded Apiospora species isolated from marine substrates in Korea with eight new combinations (A. piptatheri and A. rasikravindrae). Mycobiology. 2022;50(1):46–54.
- Wang H, Lun Y, Lu Q, et al. Eight new Arthrinium species from China. MycoKeys. 2018;39:1–27.
- Ellis MB. Dematiaceous hyphomycetes. XI. Mycologic Pap. 1972;131:1–25.
- Samuels G, McKenzie E, Buchanan DE. Ascomycetes of New Zealand 3. Two new species of Apiospora and their Arthrinium anamorphs on bamboo. New Zealand J Bot. 1981;19(2):137–149.
- Tian X, Karunarathna SC, Mapook A, et al. One new species and two new host records of Apiospora from bamboo and maize in Northern Thailand with thirteen new combinations. Life. 2021;11(10):1071.
- Janssen JJ. Mechanical properties of bamboo. Berlin (Germany): Springer Science & Business Media. 2012.
- Dai D-Q, Tang L-Z, Wang H-B. A review of bambusicolous ascomycetes. Bamboo: Curr Future Prosp. 2018;165(10):5772.
- Hyde K, Zhou D, Dalisay T. Bambusicolous fungi: a review. Fungal Divers. 2002;9:1–14.
- Helander M, Jia R, Huitu O, et al. Endophytic fungi and silica content of different bamboo species in giant panda diet. Symbiosis. 2013;61(1):13–22.
- Kim J-J, Lee S-S, Ra J-B, et al. Fungi associated with bamboo and their decay capabilities. Holzforschung. 2011;65(2):271–275.
- Lee KS, Jung SY, Son YM, et al. Biomass estimation of Phyllostachys pubescens stands in KFRI, Southern Forest Research Center. J Kor Soc Forest Sci. 2012;101(1):138–147.
- Park H, Lee J-C, Eom A-H. First reports of five endophytic fungi isolated from leaves of plants inhabiting the Hansando Island in Korea. Kor J Mycol. 2020;48(3):217–228.
- Das K, Lee S-Y, Choi H-W, et al. Taxonomy of Arthrinium minutisporum sp. nov., Pezicula neosporulosa, and Acrocalymma pterocarpi: new records from soil in Korea. Mycobiology. 2020;48(6):450–463.
- White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, et al. editors. PCR protocols: a guide to methods and applications. New York (NY): Academic Press; 1990. p. 315–322.
- Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes‐application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2(2):113–118.
- Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172(8):4238–4246.
- Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91(3):553–556.
- Rehner SA, Buckley E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 2005;97(1):84–98.
- Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 1995;61(4):1323–1330.
- O’Donnell K, Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol. 1997;7(1):103–116.
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780.
- Katoh K, Misawa K, Kuma K-I, et al. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–3066.
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313.
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755.
- Darriba D, Taboada GL, Doallo R, et al. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9(8):772.
- Senanayake IC, Maharachchikumbura SS, Hyde KD, et al. Towards unraveling relationships in Xylariomycetidae (Sordariomycetes). Fungal Divers. 2015;73(1):73–144.
- Pintos Á, Alvarado P, Planas J, et al. Six new species of Arthrinium from Europe and notes about A. caricicola and other species found in Carex spp. hosts. MycoKeys. 2019;49:15–48.
- Dai DQ, Phookamsak R, Wijayawardene NN, et al. Bambusicolous fungi. Fungal Divers. 2017;82(1):1–105.
- Crous PW, Hernández-Restrepo M, Schumacher R, et al. New and interesting fungi. 4. Fungal Syst Evol. 2021;7:255–343.
- Hyde KD, Hongsanan S, Jeewon R, et al. Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016;80(1):1–270.
- Senanayake IC, Bhat JD, Cheewangkoon R, et al. Bambusicolous Arthrinium species in Guangdong province, China. Front Microbiol. 2020;11:602773.
- Tang X, Goonasekara ID, Jayawardena RS, et al. Arthrinium bambusicola (Fungi, Sordariomycetes), a new species from Schizostachyum brachycladum in Northern Thailand. Biodivers Data J. 2020;8:e58755.
- Feng Y, Liu J-KJ, Lin C-G, et al. Additions to the genus Arthrinium (Apiosporaceae) from bamboos in China. Front Microbiol. 2021;12:661281.
- Jayasiri SC, Hyde KD, Ariyawansa HA, et al. The faces of fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 2015;74(1):3–18.
- Dai D-Q, Jiang H-B, Tang L-Z, et al. Two new species of Arthrinium (Apiosporaceae, Xylariales) associated with bamboo from Yunnan, China. Mycosphere. 2016;7(9):1332–1345.
- Jiang N, Liang YM, Tian CM. A novel bambusicolous fungus from China, Arthrinium chinense (Xylariales). Sydowia. 2020;72:77–83.
- Hyde KD. Refined families of Sordariomycetes. Mycosphere. 2020;11(1):305–1059.
- Bhunjun CS, Niskanen T, Suwannarach N, et al. The numbers of fungi: are the most speciose genera truly diverse? Fungal Divers. 2022;114(1):387–462.
- Yang C-L, Xu X-L, Dong W, et al. Introducing Arthrinium phyllostachium sp. nov. (Apiosporaceae, Xylariales) on Phyllostachys heteroclada from Sichuan province, China. Phytotaxa. 2019;406(2):91–110.
- Jiang N, Li J, Tian C. Arthrinium species associated with bamboo and reed plants in China. Fungal Syst Evol. 2018;2(1):1–9.
- Majeedano AQ, Chen J, Zhu T, et al. The first whole genome sequence discovery of the devastating fungus Arthrinium rasikravindrae. J Fungi. 2022;8(3):255.
- Hino I, Katumoto K. Illustrations fungorum bambusicolorum VIII. Bull Fac Agric Yamaguchi Univ. 1960;11:9–34.
