Abstract
A powdery mildew (Erysiphaceae) has been continuously collected on the leaves of Lonicera harae in the southern part of the Korean Peninsula, where this shrub is indigenous. Microscopic examination of the asexual morphs revealed that the current collections are differentiated from the all known Erysiphe species on Lonicera spp. by its longer conidiophores and longer conidia. Although the morphology of the chasmothecia is reminiscent of Erysiphe ehrenbergii and E. lonicerae, the specimens on L. harae differ from them in having smaller ascospores. A phylogenetic tree generated from a combined dataset of the internal transcribed spacer region and 28S rDNA gene sequences demonstrates that sequences obtained from three powdery mildew collections on L. harae clustered together as an independent species clade with high bootstrap values distant from other Erysiphe species on Lonicera, representing a species of its own. Based on morphological differences and molecular-phylogenetic results, the powdery mildew on L. harae is proposed as a new species, Erysiphe lonicerigena, and the holomorph of the fungus is described and illustrated in this study.
1. Introduction
The genus Lonicera L., commonly known as honeysuckles dominates within the family Caprifoliaceae by having almost 180 deciduous plant species, which are planted for their ornamental value in countries of the Northern Hemisphere [Citation1]. In Korea, 17 indigenous species (including one variety) and more than 20 species/varieties are listed within this genus (http://www.nature.go.kr/kpni/index.do). Many phytopathogenic fungi associated with Lonicera species have been recorded in Korea [Citation2].
Among phytopathogenic fungi associated with various plant species of Lonicera, powdery mildews from the genus Erysiphe (Helotiales, Ascomycota) have been recorded on 39 Lonicera species globally [Citation3], whereas eight of them are so far known from Korea [Citation4–6]. Shin [Citation7] listed five species of Lonicera in Korea as host plants of Microsphaera erlangshanensis Y.N. Yu. As stated in the latest studies on Erysiphe lonicerae complex, Erysiphe erlangshanensis (Y.N. Yu) U. Braun & S. Takam. (syn. M. erlangshanensis) is currently known as pathogen of Lonicera chrysantha Turcz. ex Ledeb., L. maackii (Rupr.) Maxim., L. subsessilis Rehder, L. tatarica L., and L. vidalii Franch. & Sav. in Korea, while E. lonicerae DC. is confined to L. japonica Thunb., L. sempervirens L. and L. periclymenum L. [Citation3].
During our extensive studies around the Korean peninsula, leaves of Lonicera harae Makino affected with an unknown Erysiphe sp. were continuously collected from the southern part of Korea (). To date, no powdery mildew species was reported on L. harae globally. Therefore, this study aims at describing and illustrating the powdery mildew found on L. harae in Korea, and at clarifying its phylogenetic position within the Erysiphe-Lonicera complex within the scope of a taxonomic revision of this association globally.
Figure 1. (A) Young shoots of Lonicera harae infected by Erysiphe lonicerigena. (B) Early symptoms appeared on rachis and leaflets in spring. (C) Symptoms appeared on leaflets in autumn. (D) Chasmothecia formed on the upper leaf surface. (E) Close-up view of chasmothecia. (F–H) Conidiophores. (I) Conidia. (J,K) Chasmothecia producing several asci. (L,M) Chasmothecial appendages. (N) Asci containing 5–6 ascospores each.

2. Materials and methods
2.1. Specimen collection
Seven specimens have been deposited in the mycological herbarium of the Korea University (KUS-F), Seoul, Korea. Since L. harae is distributed in the southern part of the Korean Peninsula in Korea [Citation8], all specimens were collected in 2022, when our extensive survey for phytopathogenic fungi was performed in Jeollabukdo province. Detailed information for these specimens is listed in the Taxonomy section.
2.2. Morphological observations
Detailed morphological characteristics of the fungus were examined using fresh materials. The mycelial felt was scraped from the leaves, mounted in a drop of sterile water and then examined under an optical microscope (Zeiss AX10 microscope equipped with an AxioCam MRc5, Carl Zeiss, Germany). To examine the sexual morphs, chasmothecia were picked up with a sterile needle and mounted in 3% NaOH solution. Each morphological structure was taken from at least 20 measurements.
2.3. Molecular-Phylogenetic analyses
The following three specimens were involved in the molecular-phylogenetic analyses, viz., KUS-F32778, F32811, and F32838. A whole-cell of DNA was extracted from mycelia using MaglistoTM 5 M plant genomic DNA extraction kit (Bioneer, Daejeon, Korea) according to the manufacturer’s protocol. The nucleotide sequences of ITS1–5.8–ITS2S (internal transcribed spacer) and 5′-end of 28S rDNA gene (large subunit gene; LSU) were determined with the application of primer pairs ITS1/PM6 and PM3/NLP2, respectively [Citation9]. Obtained sequences of forward and reverse were assembled using BioEdit 7.2 software [Citation10] and the resulting amplicons were deposited into the GenBank under the Accession Nos: OQ221136 and OQ221120 for ITS, and OQ221121, OQ221127, and OQ221128 for LSU. For phylogenetic analyses, a combined dataset of ITS and LSU was created in MEGA 11 [Citation11] using three newly obtained and 35 closely related sequences of the genus Erysiphe retrieved from GenBank. Two sequences of Erysiphe corylopsidis Shiroya & S. Takam. (AB478988, LC270849) were selected as outgroups. Phylogenetic trees were generated by the maximum parsimony (MP) and maximum likelihood (ML) methods in PAUP* 4.0a and MEGA11, respectively [Citation11,Citation12]. The strength of the internal branches in the resulting tree was tested from 1000 replications in bootstrap (BS) analysis. BS values higher than 70% are shown on the related branches. The tree scores, such as tree length, consistency index, retention index, and rescaled consistency index (RC) were calculated.
3. Results
3.1. Taxonomy
Erysiphe lonicerigena I.Y. Choi, L. Abasova & H.D. Shin, sp. nov. ()
MycoBank no: MB848311
Etymology: The specific epithet was derived from the generic name of the host plant and the Latin suffix -gena (= born from).
Diagnosis: Morphologically close to but phylogenetically distinct from Erysiphe ehrenbergii and E. lonicerae s. str.; the asexual morph differs from the latter two species in having longer conidiophores (74–136(–210) µm) and longer conidia (34–58 µm); the sexual morph differs from them in having shorter chasmothecial appendages, 1.0–1.5 times (vs. 1.0–2.5 times and 1.0–3.0 times, respectively).
Typification: on L. harae, KOREA, Wanju, Daea Arboretum, 2 May 2022, leg. and det. H.D. Shin, KUS-F32778 (holotype), LSU sequence OQ221128; KOREA, Imsil, Saseondae Park, 17 November 2022, leg. and det. H.D. Shin, KUS-F33567 (paratype).
Description: Mycelium amphigenous, sometimes abundantly epiphyllous, thin, effuse, covering the whole leaf surface. Hyphal appressoria moderately lobed to nipple-shaped, positioned in pairs or solitary, and 4–9 µm wide. Conidiophores mostly single, occasionally two or three on a mother cell, straight or slightly flexuous at the base of foot-cells, 74–136(–210) × 8–9 µm, and producing conidia singly. Conidia cylindrical to cylindrical-oval, devoid of conspicuous fibrosin bodies, 34–58 × 17–20 µm with a length/width ratio of 1.9–3.0, producing germ tubes on the perihilar position of the conidia. Chasmothecia dark brown, spherical, scattered to subgregarious, and 92–110 µm in diameter. Appendages equatorial, straight, not stiff, 3–5 times dichotomously branched, cover, aseptate or 1-septate, 10–16 per chasmothecium, erect, 1.0–1.5 times as long as the chasmothecial diam., 6–9 µm wide at the base, becoming narrower toward the tip, secondary and/or third branches elongated, ultimate tips mostly recurved, occasionally straight, and brown at the base, becoming paler toward the tip. Asci 4–7 per chasmothecium, short-stalked or subsessile, 5–6-spored, and 45–54 × 32–42 µm. Ascospores subhyaline, ellipsoid-ovoid, and 14–18 × 10–12 µm with a length/width ratio of 1.3–1.8.
Additional materials examined: on L. harae, KOREA, Wanju, near Songkwangsa Temple, 9 May 2022, KUS-F32802; Wanju, Sangkwan Forest, 11 May 2022, KUS-F32811; Jeongeup, Naejangsan National Park, 20 May 2022, KUS-F32838; Jeongeup, near Museongseowon Confucian Academy, 20 May 2022, KUS-F32844; Wanju, Daea Arboretum, 3 Jun 2022, KUS-F32898.
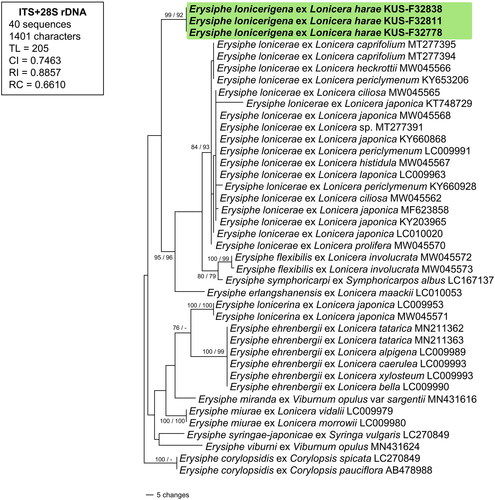
Notes: The current specimens of Erysiphe collected on L. harae are morphologically differentiated from all related species of Erysiphe. Although the conidiophores and conidia of Erysiphe lonicerina S. Takam. known on Lonicera japonica from Japan, are more or less similar to those of the current species, the sexual stage of E. lonicerina is so far not known [Citation3]. In the phylogenetic tree, E. lonicerina is clustered in a separate clade apart from E. lonicerigena, which confirms that these two species being distinct from each other (). This latter species is characterized by having longer conidiophores and longer conidia compared to other Erysiphe spp. known on Lonicera spp. Despite the morphological similarity in the characteristics of chasmothecia between E. ehrenbergii M. Bradshaw & S. Takam., E. lonicerae s. str. and E. lonicerigena, our new species differs from the first two species in having much smaller ascospores, 14–18 × 10–12 µm (vs. 14–26 × 9–14 µm and 15–24 × 8–15 µm, respectively).
Figure 2. A phylogenetic tree of Erysiphe spp. on Lonicera spp. generated from a combined dataset of ITS and LSU from 40 Erysiphe sequences. The representing tree was constructed based on the MP method in PAUP* 4.0a. The resulting sequences obtained in this study were shown in boldface. BS values higher than 70% obtained from MP and ML analyses were given on the related branches, respectively.
3.2. Molecular phylogeny
Newly obtained two sequences of ITS (552, 553 bp) and three for LSU (854, 855 and 962 bp) were compared with reference sequences in GenBank using BLASTn search tool. Results for ITS showed 97.08% similarity with sequences of E. syringae-japonicae (LC270849, AB295458) and 96.9% with E. lonicerae (KY660928, KY660907, etc), followed by similar results of LSU, which were 98.7% resemble to Erysiphe erlangshanensis (LC010053), 98.6% E. pseudocorylacearum (LC009928) and etc. The data matrix consists of 40 sequences and 1401 characters, of which 45 (3.2%) were variable and 87 (6.2%) were informative for parsimony analysis. Phylogenetic trees generated from ML and MP analyses were almost identical to each other. In the given MP tree (), isolates of E. lonicerigena on L. harae were clustered in a separate clade from all other Erysiphe spp. on Lonicera spp. with support of 99% and 92% BS values obtained from MP and ML analyses, respectively.
4. Discussion
In the monograph of the Erysiphales, six powdery mildews species from the genus Erysiphe section Microsphaera have been described on Lonicera species globally [Citation13]. Recently published taxonomic revisions of the Erysiphe-Lonicera complex were accomplished by Bradshaw et al. based on morphological and molecular-phylogenetic approaches [Citation3]. In that study, the number of Erysiphe species on Lonicera spp. was increased to eight, of them E. ehrenbergii (China, Japan), E. erlangshanensis (China, Japan, Korea), E. lonicera (China, Japan, Korea) and E. lonicerina (Japan) were reported from East Asian countries [Citation14]. As stated by Bradshaw et al. [Citation3], much more collections assigned to E. lonicerae s. lat. and Microsphaera penicillata s. lat. on Lonicera spp. should be studied. The new species proposed in this study is morphologically and phylogenetically clearly distinct from all known Erysiphe species on Lonicera.
Lonicera harae belongs to the subgenus Chamaecerasus section lsica, phylogenetically positioned with L. fragrantissima and L. hispida in a single clade and is distributed in the southern part of Korean peninsula, Jeju Island (Korea), Tsushima island (Japan), and some south-eastern part of China [Citation15]. It is an open question whether E. lonicerigena is confined to L. harae or not. According to summarized information on host range based on the subgenus level outlined in Bradshaw et al., E. ehrenbergii, E. erlangshanensis, E. flexibilis M. Bradshaw, U. Braun & S. Takam., E. lonicera-ramossimae (Tanda) U. Braun, S. Takam and E. miurae (U. Braun) U. Braun & S. Takam are associated with host species from the subgenus Chamaecerasus, whereas hosts of E. lonicerae and E. lonicerina belong to the subgenus Lonicera [Citation3,Citation15]. Strict host speciation occurred in most of Erysiphe spp. on Lonicera. However, E. magnusii (S. Blumer) U. Braun and S. Takam. represents an exception, since most of the host plants of this fungus are from the subgenus Lonicera and some of them are from the subgenus Chamaecerasus. Nevertheless, the phylogenetic tree does not represent accuracy on host speciation at the subgenus level. Thereby, powdery mildew species on Lonicera spp. from both subgenera are scattered, and do not make a distinct group based on the subgenus level. However, it is assumed that each powdery mildew species evolved with host species from one subgenus.
Conclusively, this is the first report on the occurrence of a powdery mildew on L. harae globally. With this work, the diversity of Erysiphe on Lonicera spp. has been expanded into nine distinct species.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Mabberley DJ. Mabberley’s plant-book: a portable dictionary of plants. 3rd ed. Avon: Cambridge University Press, 2008.
- Korean Society of Plant Pathology. List of plant diseases in Korea. 6th ed. Seoul, Korea: Korean Society of Plant Pathology; 2022.
- Bradshaw M, Braun U, Gotz M, et al. Taxonomy and phylogeny of the Erysiphe lonicerae complex (Helotiales, Erysiphaceae) on Lonicera spp. Fungal Syst Evol. 2021;7(1):49–65.
- Kim KH, Lee SK, Shin HD. First report of powdery mildew caused by Erysiphe lonicerae var. lonicerae on Lonicera sempervirens in Korea. Plant Pathol. 2008;57(2):374–374.
- Lee SH, Lee CK, Cho SE, et al. First report of powdery mildew caused by Erysiphe lonicerae var. lonicerae on Lonicera japonica in Korea. Plant Dis. 2016;100(4):856–856.
- Choi IY, Abasova L, Choi JH, et al. First report of powdery mildew caused by Erysiphe lonicerae on Lonicera periclymenum in Korea. J Plant Pathol. 2022;104(3):1153–1153.
- Shin HD. Erysiphaceae of Korea. Suwon, Korea: National Institute of Agricultural Science and Technology; 2000.
- Son YH, Park SH, Jeong DH, et al. Growing environment characteristics and vegetation structure of Lonicera harae, medicinal plant. Korean J. Plant Res. 2021;34:297–310.
- Bradshaw M, Tobin PC. Sequencing herbarium specimens of a common detrimental plant disease (powdery mildew). Phytopathology. 2020;110(7):1248–1254.
- Hall TA. BioEdit: a User-Friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98.
- Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38(7):3022–3027.
- Swafford DL. PAUP: phylogenetic analyses using parsimony (and other methods) 4.0b10. Sunderland, MA: Sinauer; 2002.
- Braun U, Cook RTA. Taxonomic manual of the Erysiphales (powdery mildews). CBS biodiversity series. Utrecht, the Netherlands: CBS-KNAW Fungal Biodiversity Centre; 2012.
- Farr DF, Rossman AY. Fungal Databases, Systematic Mycology & Microbiology Laboratory, ARS, USDA. Available from: http://nt.ars-grin.gov/fungaldatabases/. Accessed 3 January 2023.
- Nakaji M, Tanaka N, Sugawara T. A molecular phylogenetic study of Lonicera L. (Caprifoliaceae) in Japan based on chloroplast DNA sequences. Acta Phytotaxonomica Geobotanica. 2015;6:137–151.
