Abstract
In this study, a fungal strain KNUF-22-025 belonging to the genus Botryotrichum was isolated from the soil in Korea. The cultural and morphological characteristics of this strain differed from those of closely related species. On malt extract agar, strain KNUF-22-025 showed slower growth than most of the related species, except B. domesticum. The conidia size (9.6–21.1 × 9.9–18.4 µm) of strain KNUF-22-025 was larger than those of B. piluliferum, B. domesticum, and B. peruvianum but smaller than those of B. atrogriseum and B. iranicum. Conidiophores in strain KNUF-22-025 (137 µm) were longer than those in other closely related species but shorter than those in B. atrogriseum. Multi-locus analysis of molecular markers, such as ITS, 28S ribosomal DNA, RBP2, and TUB2 revealed that strain KNUF-22-025 was distinct from other Botryotrichum species. Thus, this strain is proposed as a novel species based on morphological characteristics along with molecular phylogeny and named Botryotrichum luteum sp. nov.
1. Introduction
Sordariomycetes is the second largest group in the phylum Ascomycota and has considerable importance [Citation1]. Currently, 47 orders, 127 families, 1975 genera, and 23,187 species have been recognized in this phylum [Citation2]. Fungi of this phylum span various ecologies, including mycoparasites, animal and plant pathogens, and endophytes [Citation3]. The order Sordariales is a diverse group of fungi within the class Sordariomycetes. Numerous saprotrophic fungi found in soil, plant materials, and dung from wild animals are included in this group [Citation4]. Winter (1885) established the family Chaetomiaceae, which is characterized by Chaetomium, and the family Chaetomiaceae is included in Sordariales, which comprises 25 genera [Citation3–8]. The Chaetomiaceae, genus Botryotrichum is characterized by thick-walled conidia with setose, branching, and macronematous conidiophores. Owing to the sexual state of some Botryotrichum species, particularly that of Botryotrichum piluliferum (Type strain), which was previously regarded as Chaetomium, a connection between the two genera was discovered [Citation8,Citation9]. Daniels (1961) collected the B. piluliferum strain from cellulose films that were incubated in soil. The collected strain showed unbranched ascomatal hairs with circinate tips and ellipsoid ascospores, which are the characteristics of Chaetomium murorum. As a result, Chaetomium piluliferum was assigned a new holomorphic name [Citation9]. Wang et al. (2016), demonstrated that C. murorum, Emilmuelleria spirotricha, and Thermomyces verrucosus were related to Botryotrichum species, and new combinations, such as B. murorum, B. spirotrichum, and B. verrucosum, were proposed based on phylogenetic analyses [Citation8,Citation10]. Currently, 23 species in Botryotrichum have been listed in the Index Fungorum (www.indexfungurum.org). Of the species of Botryotrichum, seven are anamorphically characterized, including B. piluliferum, B. atrogriseum, B. verrucosum, B. peruvianum, B. domesticum, B. foricae, and B. iranicum [Citation8–12]. Recently in 2022, new species of B. inquinatum, B. retardatum, B. trichorobustum, B. vitellinum, and B. geniculatum were included in Botryotrichum [Citation13].
This study aimed to explore and classify the novel species of fungi isolated from soil in Korea. The novelty of the strain was confirmed via cultural, morphological, and molecular characteristics. In this study, a new species of fungus was discovered in Korea and given a comprehensive description as well as an illustration.
2. Materials and methods
2.1. Sample collection and isolation of fungal strain
The soil sample was collected from Subuksan, Okcheon-gun, Chungcheongbuk-do (36°23′45″N, 127°39′19″E), Korea. Isolation of the strain was performed using the standard serial dilution technique [Citation14]. Sample soil (1 g) was added to 10 Ml of sterile distilled water, and the soil suspension was diluted serially and spread on potato dextrose agar plates (PDA; Difco, Detroit, MI). The plates were incubated for 3–4 days at 25 °C until single colonies were observed. Further, single colonies were transferred to new PDA plates and incubated at 25 °C. The strain KNUF-22-025 was selected based on cultural, morphological, and phylogenetic analyses.
2.2. Morphological characterization of strain KNUF-22-025
The cultural and morphological characteristics of the strain KNUF-22-025 were observed using four different media viz. PDA, oatmeal agar (OA), malt extract agar (MEA), and potato carrot agar (PCA) [Citation15]. The colony characteristics of the strain were recorded, and the mycological characteristics were observed using a light microscope (BX-50; Olympus, Tokyo, Japan).
2.3. Genomic DNA extraction, PCR amplification, and sequencing
The total genomic DNA from fungal mycelia of the strain KNUF-22-025 was extracted using a HiGene Genomic DNA prep kit (BIOFACT, Daejeon, Korea) according to the manufacturer’s instructions. The following combined datasets were used according to previous studies: internal transcribed spacer (ITS) regions, 28S ribosomal DNA (LSU), the second largest subunit of RNA polymerase II (RPB2) genes, and β-tubulin (TUB2) [Citation10,Citation12]. The primer pairs of ITS1F/ITS4, NL1/NL4, RPB2AM-1gf/RPB2AM-7R, and T1/T2 were used for ITS, LSU, RPB2, and TUB2, respectively [Citation16–18]. The thermal conditions for PCR amplification were used as described in previous studies [Citation12, Citation19]. PCR products were confirmed by running the amplified DNA samples in 1.2% agarose gel stained with ethidium bromide. The purification of the PCR products was determined using EXOSAP-IT (Thermo Fisher Scientific, Waltham, MA, USA) and sequenced at Solgent Co., Ltd., Korea. The DNA sequence data were analyzed using SeqMan Lasergene software (DNAStar Inc., Madison, Wisconsin, USA).
2.4. Phylogenetic analyses
Using the Basic Local Alignment Search Tool (BLAST), four molecular markers (ITS, LSU, RPB2, and TUB2) of strain KNUF-22-025 were compared with reference sequences from the GenBank database of the National Center for Biotechnology Information (NCBI) (). Based on Kimura’s neighbor-joining algorithm, the evolutionary distance matrices were generated [Citation20]. To determine the precise taxonomic position of the strain, neighbor-joining [Citation21], maximum likelihood [Citation22], and maximum parsimony [Citation23] trees were constructed. The MEGA 7.0 software program was used for phylogenetic tree analysis, and bootstrap values were based on 1000 replications [Citation24].
Table 1. GenBank accession numbers of fungal strains used in this study for the phylogenetic analysis.
3. Results
3.1. Taxonomical analysis of Botryotrichum luteum sp. nov.
Strain KNUF-22-025 was identified as a novel species based on differences in morphological and molecular characteristics from closely related Botryotrichum species.
Botryotrichum luteum J. J. Ryu, S.Y. Lee, and H.Y. Jung, sp. nov. ()
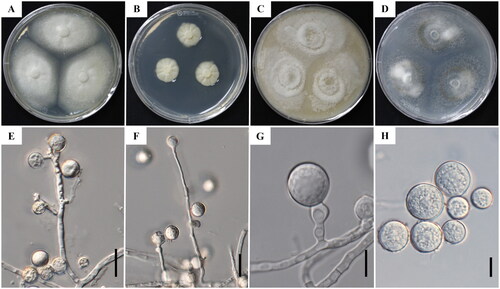
Figure 1. Cultural and morphological characteristics of Botryotrichum luteum (KNUF-22-025T). Colonies on potato dextrose agar (A), malt extract agar (B), oatmeal agar (C), and potato carrot agar (D), after 14 days incubation at 25 °C. Conidiophores and conidiogenous cells (E-G); Conidia (H). Scale bars: E, F = 20 µm; G, H = 10 µm.
MycoBank: MB 845982
Etymology: The specific name “luteum” represents yellow color colonies.
Typus: Isolated from mountain soil containing plant debris from Subuksan, Okcheon-gun, North Chungcheong Province (36°23′45″N, 127°39′19″E), Korea. The metabolically inactive stock culture of the strain (NIBRFGC000509835) was deposited at the culture center of National Institute of Biological Resources (NIBR).
Ecology and distribution: Several species have been isolated from mountains and field soils, the dung of animals such as rabbits and deer, etc. Additionally, members of this genus were isolated from the air and indoor environments, such as inside villas and ceiling tiles. In the present study, the proposed novel species Botryotrichum luteum was isolated from the forest soil in South Korea.
Cultural characteristics: The colonies were smooth, round, and flat with lobate margins spreading moderate aerial mycelium. Colonies were 63.5–66.3 mm in diameter after culturing for 14 days at 25 °C and the reverse colonies were yellow to light beige on PDA (). On MEA, the colonies grew very slowly, the surface produced floccose white colonies, and attained 21.5–24.0 mm in diameter within 14 days of growth at 25 °C with reverse pale yellow to light beige (). On OA media, the colony surface was floccose to funiculose, and the mycelium color was white with dark brown to black spots. Mycelium diameter was 74.5–80.0 mm after 14 days of growth at 25 °C (). On PCA, the fungal colonies were white to dirty white with 81.5–83.0 mm diameter after a growth period of 14 days at 25 °C. The color of the reverse colonies was pale yellow to light beige ().
Morphological characteristics: The conidiophores were produced laterally from hyphae near the base with a diameter of 2.5–5.3 µm and a length of upto 137 µm. The mycelium was immersed, colorless, and 1.8–2.2 µm thick (). Conidiophores were micronematous, present on both submerged and aerial hyphae, reduced to conidiogenous cells when multiple-celled, unbranched or branching, and colorless to light brown. Conidiogenous cells were solitary, monoblastic, hyaline, smooth, terminal, intercalary, or occasionally integrated into the hyphae. The cells were barrel-shaped, cylindrical, or T-shaped (). Conidia were solitary, partial hyaline when young, one-celled, globose to subglobose, rarely in chains of coupled spores, smooth to slightly roughened, 9.6–21.1 × 9.9–18.4 µm (x̅ = 15.5 × 14.6 µm, n = 50) with a length/width ratio of 1.05 and visible hilum, 3–6.4 µm wide, and 2.7–4.2 µm high ().
Note: B. piluliferum, B. atrogriseum, B. domesticum, B. iranicum, B. peruvianum, and B. verrucosum are all phylogenetically related to the strain KNUF-22-025. However, several morphological characteristics distinguished it from related species (). Strain KNUF-22-025 achieved a growth of 21.5-24.0 mm in diameter after culturing for 14 days at 25 °C on MEA, whereas the related strain B. domesticum (UAMH 11929T- TType strain). Grew 5.5 mm in diameter in 1 month at 25 °C. The other related species, such as B. iranicum (ABRIICC 10152T), B. piluliferum (DTO 254-B8), B. peruvianum (CBS 421.93), and B. verrucosum (CBS 116.64T), showed growths of 22–23, 25–31, 21–28, and 17–23 mm, respectively, after 7 days at 25 °C (). The strain KNUF-22-025 produced white colonies with reverse pale yellow to light beige, whereas B. iranicum displayed white to pinkish buff, reverse light ochraceous-buff. The conidia produced by KNUF-22-025 were larger in size than those produced by B. piluliferum (11.0–17.5 µm), B. domesticum (15.1–20.3 14.8–19.8 µm), B. peruvianum (12.0–16.0 µm), and B. verrucosum (14.0–15.5 µm) and smaller than those produced by B. atrogriseum (10.0–25.0 µm) and B. iranicum (9.5–23.5 × 9.5–17.0 µm) (). The B. iranicum and B. atrogriseum strains closest to strain KNUF-22-025 produced conidia with rough and verrucose surfaces, respectively (). However, the conidia of strain KNUF-22-025 were mostly smooth with a slight rough surface. The conidiophore of KNUF-22-025 (137 µm) was longer than those of B. piluliferum (40 µm), B. iranicum (70 µm), B. verrucosum (65 µm), and B. peruvianum (60 µm) but shorter than those of B. atrogriseum (150 µm). Thus, strain KNUF-22-025 is well distinguished from the previously described closest Botryotrichum species.
Table 2. Morphological characteristics of Botryotrichum luteum (KNUF-22-025T) and comparison with closely related species grown on MEA.
3.2. Molecular phylogeny of strain KNUF-22-025
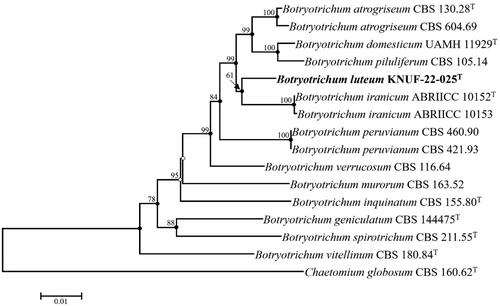
The obtained lengths of the sequences were 583, 576, 627, and 642 bp for ITS, LSU, RPB2, and TUB2, respectively. These sequences were deposited in NCBI GenBank with accession numbers LC731694, LC731695, LC731696, and LC731697 for ITS, LSU, RPB2, and TUB2, accordingly. Strain KNUF-22-025 was found to be 99.83% similar to B. piluliferum, (CBS 503.83), 99.82% to B. atrogriseum (CCF 5752), and 99.49% to B. domesticum (UAMH 11929T) in the ITS regions. Additionally, in the LSU, it showed 100% similarities to B. piluliferum (CBS 503.83), and B. verrucosum (CBS 116.64T) and 99.83%, and 99.65% to B. atrogriseum (CCF 5752) and B. domesticum (UAMH 11929T), respectively. Similarly, RPB2 gene sequence analysis of KNUF-22-025 showed 97.28%, 96.55%, 95.69%, 95.59%, and 95.40% similarities to B. verrucosum (CBS 116.64), B. atrogriseum (CBS 604.69, CBS 130.28T), B. domesticum (UAMH 11929T), B. peruvianum (CBS 460.90), and B. piluliferum (DTO 254-B9), respectively. Moreover, TUB2 gene sequences of KNUF-22-025 showed 94.58% and 94.28% similarities with B. domesticum (UAMH 11929T) and B. piluliferum (CBS 654.79), respectively. Also B. atrogriseum (CBS 130.28T, CCF 5752, and CBS 604.69) displayed 95.47 to 96.14% similarities based on TUB2 gene. Additionally, ITS, LSU, and TUB2 sequences of strain KNUF-22-025 showed a high similarity to B. foricae with 99.55%, 99.83%, and 94.28% similarities, respectively. Although ITS and LSU sequences of strain KNUF-22-025 showed a high similarity (99.8%–100%) with the strain B. iranicum, TUB2 and RPB2 sequences of this strain depicted a high similarity to B. iranicum (94.94%) and B. atrogriseum (96.14%, 96.55%), respectively. For more precise identification, we performed multilocus sequence analysis using the combined dataset sequences of the ITS regions, LSU, RPB2, and TUB2 genes of strain KNUF-22-025. The combination of these four molecular markers has shown to be highly effective in the identification of species using neighbor-joining (NJ), maximum parsimony (MP), and maximum likelihood (ML) phylogenetic trees. The neighbor-joining phylogenetic tree node as well as the filled nodes in the maximum parsimony and maximum likelihood trees were used to construct the phylogenetic analysis using the combined dataset of the ITS regions, LSU, RPB2, and TUB2 (). As indicated in the open circles, the corresponding nodes were also constructed using the maximum likelihood or maximum parsimony algorithms. The precise taxonomic position of the strain was ascertained by a phylogenetic analysis based on a combination of sequences with maximum parsimony (tree length =733, consistency index = 0.58, retention index = 0.62, and composite index = 0.45). The results of the phylogenetic tree demonstrated that strain KNUF-22-025 did not cluster with closely related fungal strains. Hence, the strain KNUF-22-025 is proposed as a novel species within the Botryotrichum genus.
Figure 2. Neighbor-Joining phylogenetic tree based on molecular markers the internal transcribed spacer (ITS) regions, 28S ribosomal DNA (LSU), the second largest subunit of RNA polymerase II (RPB2), and β-tubulin (TUB2) genes sequences showing the phylogenetic position of the strain KNUF-22-025. Chaetomium globosum CBS 160.62T comprised the outgroup. The neighbor-joining phylogenetic tree, maximum likelihood, and maximum parsimony trees indicated with filled nodes, whereas open circles showed maximum-likelihood or maximum-parsimony. The numbers (>60%) above the branches represent the bootstrap values obtained for 1,000 replicates. The isolated strain is shown in bold. Bar, 0.01 substitutions per nucleotide position.
4. Discussion
Previous studies have reported that the strains belonging to the genus Botryotrichum were widely discovered in the excrement of various animals, including rabbit, herbivore, deer, donkey, and field soil [Citation10]. The strains B. piluliferum and B. murorum have been discovered from interior substrates, such as plaster walls, villa walls, ceiling tiles, and anthropogenic environments, whereas B. peruvianum strain has been found in indoor air. The strain B. murorum was identified in the liquor cerebrospinalis of Homo sapiens [Citation10]. Secondary metabolites produced by fungi are well-known for their various biological properties, which led to the development of uncounted agrochemicals and life-saving medicines. B. piluliferum, the mycotoxin-producing type species of Botryotrichum, produces oxisterigmatocystins E, G, and H, which have shown antimalarial activity against Plasmodium falciparum and cytotoxic activity against KB, MCF-7, and NCI-H187 cell lines [Citation27]. These two strains also produce altersolanol C, diorcinol, and botryobutenolide A, which have shown antibacterial activity against Staphylococcus aureus and Enterococcus faecalis [Citation28]. Additionally, the strain of B. piluliferum has a wide ecological distribution and has been identified as a chili pepper seed-borne fungus [Citation29]. B. murorum has previously been recognized as a human pathogen that causes phaeohyphomycosis [Citation30]. Additionally, strains B. atrogriseum and B. iranicum (Type strain) were isolated from the cornfield soil in Canada and Iran, respectively [Citation19,Citation25]. In Korea, Botryotrichum sp. was identified in the upland soil and cultivated garlic in Chungcheongbuk-do, Korea [Citation31]. Botryotrichum sp., along with several other fungi and bacteria, was identified from Nuruk, a Korean cereal fermentation starter [Citation32]. Recently, B. iranicum was identified in soil samples from Korea [Citation33]. So, there are several species of Botryotrichum that produce mycotoxins, and metabolites, show anti-malarial activities, and the genus has a wide range of distribution, and pathogenicity which is not been explored thoroughly even in Korea. As an outcome, a novel species (B. luteum) was discovered in Korean soil during this current investigation. Thus, the importance of B. luteum can be referred to by its potential for medicines or biological use which is needed to do further research.
In conclusion, the genus Botryotrichum is broadly distributed and possesses cytotoxic activities, therefore, systematic investigation of strains of these species obtained from natural sources is required to evaluate their bioactive potential. The findings of this study might guide future studies on this strain. Hence, further understanding of the phylogeny and ecology of Botryotrichum species is essential.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Kirk PM, Cannon PF, Minter DW, et al. Ainsworth and bisby’s dictionary of the fungi. 10th ed. Wallingford: CABI International; 2008. P. 644.
- Bánki O, Roskov Y, Döring, et al. Catalogue of life checklist. Catalogue of life; 2022.
- Maharachchikumbura SSN, Hyde KD, Jones EBG, et al. Towards a natural classification and backbone tree for Sordariomycetes. Fungal Divers. 2015;72:199–301.
- Cannon PF, Kirk PM. Fungal families of the world. Wallingford: CABI Bioscience; 2007.
- Winter G. New North American fungi. J Mycol. 1885;1:101–102.
- Lumbsch HT, Huhndorf SM. Myconet volume 14. Part one. Outline of Ascomycota—2009. Part two. Notes on Ascomycete systematics. Nos. 4751–5113. Fieldiana Life Earth Sci. 2010;1:1–64.
- Maharachchikumbura SSN, Hyde KD, Jones EG, et al. Families of Sordariomycetes. Fungal Divers. 2016;79:1–317.
- Wang XW, Yang FY, Meijer M, et al. Redefining Humicola sensu stricto and related genera in the chaetomiaceae. Stud Mycol. 2019;93:65–153.
- Daniels J. Chaetomium piluliferum sp. nov., the perfect state of Botryotrichum piluliferum. Trans Br Mycol Soc. 1961;44:79–86.
- Wang XW, Houbraken J, Groenewald JZ, et al. Diversity and taxonomy of Chaetomium and lavateum-like fungi from indoor environments. Stud Mycol. 2016;84:145–224.
- Crous PW, Carnegie AJ, Wingfield MJ, et al. Fungal planet description sheets: 868–950. Persoonia. 2019;42:291–473.
- Schultes NP, Strzalkowski N, Li DW. Botryotrichum domesticum sp. nov., a new hyphomycete from an indoor environment. Botany. 2019;97:311–319.
- Wang XW, Han PJ, Bai FY, et al. Taxonomy, phylogeny and identification of Chaetomiaceae with emphasis on thermophilic species. Stud Mycol. 2022;101:121–243.
- Park SK, Ten LN, Lee SY, et al. New recorded species in three genera of the Sordariomycetes in Korea. Mycobiology. 2017;45:64–72.
- Samson RA, Houbraken J, Thrane U, et al. Food and indoor fungi. 2nd ed. Westerdijk laboratory manual series 2. Utrecht, The Netherlands: Westerdijk Fungal Biodiversity Institute; 2019.
- White TJ, Bruns TD, Lee SB, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications. In: innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. San Diego: Academic Press; 1990. p. 315–322.
- Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2:113–118.
- Miller AN, Huhndorf SM. Multi-gene phylogenies indicate ascomal wall morphology is a better predictor of phylogenetic relationships than ascospore morphology in the Sordariales (Ascomycota, Fungi). Mol Phylogenet Evol. 2005;35:60–75.
- O’Donnell K, Cigelnik E. Two divergent intragenomic Rdna ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol. 1997;7(1):103–116.
- Alidadi A, Vala SA, Jouzani GS. Botryotrichum iranicum sp. nov. and Trematosphaeria magenta sp. nov. as two new species from Iran. Mycol Progress. 2020;19(12):1575–1586.
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111–120.
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425.
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17(6):368–376.
- Fitch WM. Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool. 1971;20(4):406–416.
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874.
- Beyma JFH. Mykologische Untersuchungen. Verhandelingen der Koninklijke Nederlandsche Akademie van Wetenschappen. 1929;26:1–29.
- Rajachan OA, Kanokmedhakul K, Soytong K, et al. Mycotoxins from the fungus Botryotrichum piluliferum. J Agric Food Chem. 2017;65(7):1337–1341.
- Ebrahim W, Ebada SS. Antimicrobial metabolites from extremophilic fungus Botryotrichum piluliferum strain WESH19. Chem Nat Compd. 2021;57(4):654–658.
- Chigoziri E, Ekefan EJ. Seed borne fungi of chili pepper (Capsicum frutescens) from pepper producing areas of Benue state, Nigeria. ABJNA. 2013;4(4):370–374.
- Hurst CJ. Dirt and disease: the ecology of soil fungi and plant fungi that are infectious for vertebrates. In: Hurst CJ, editor. Understanding terrestrial microbial communities. Switzerland: Springer; 2019. p. 289–405.
- Lee SK, Suh JS, Kim YS, et al. Studies on phytotoxin in intensively cultivated upland crops II. Population and identification of soil microorganisms in rhizosphere of upland crops. J Korean Soc Soil Sci Fert. 1987;20:179–183.
- Nam K, Lee NK, Yum EJ, et al. Change in the composition and enzyme activity of culturable lactic acid bacteria in Nuruk during fermentation at different temperatures. Korean J Food Preserv. 2015;22(6):920–925.
- Lim SK, Ten LN, Avalos-Ruiz D, et al. Isolation and identification of two unreported sordariomycetes fungi in Korea: Pestalotiopsis lavate and Botryotrichum iranicum. Kor J Mycol. 2022;50:183–194.
