Abstract
In this study, fungal strains designated as KNUF-22-14A and KNUF-22-15A were isolated from soil samples in Korea. These two strains were identified based on cultural and morphological characteristics as well as phylogenetic analyses and were found to be morphologically and phylogenetically identical. Upon their morphological comparison with closely related species, such as Tolypocladium album, T. amazonense, T. endophyticum, T. pustulatum, and T. tropicale, a difference in the size of short phialides [0.6–2.4(–9.3) × 0.8–1.4 µm] was observed. Meanwhile, these strains had larger conidia (1.2–3.0 × 1.2–3.0 µm) than T. album, T. amazonense, T. endophyticum, and T. tropicale and smaller conidia than T. pustulatum. Phylogenetic analyses using a multi-locus datasets based on ITS, LSU, and SSU showed that KNUF-22-14A and KNUF-22-15A formed a distinct cluster from previously identified Tolypocladium species. Thus, these fungal strains isolated from soil in Korea are proposed as a novel species according to their characteristics and are named Tolypocladium globosum sp. nov.
1. Introduction
Many species of the order Hypocreales (Ascomycota, Sordariomycetes) coexist with other organisms, such as plants, insects, and fungi, as pathogens, saprobes, or symbionts [Citation1]. Three species with sparsely branched conidiophores, inflated phialides, and one-celled conidia carried in slimy heads were used to establish the genus Tolypocladium in 1971 [Citation2]. Currently, 49 species of Tolypocladium are listed in the Index Fungorum (www.indexfungorum.org, accessed on 1 February 2023). The Tolypocladium genus has a wide range of habitats, host/substrate relationships, and geographical distribution. Tolypocladium species have been referred to as cosmopolitan displays because they can live in a variety of habitats and hosts, such as soil, insects, plants, lichens, and hypogeal fungi [Citation3,Citation4]. This genus was established with the type species Tolypocladium inflatum, along with T. cylindrosporum and T. geodes [Citation2], after which Cordyceps subsessilis, a sexual species, and T. inflatum, an asexual species, were also added [Citation5]. Elaphocordyceps, a sexual genus, was then introduced. Multigene phylogenetic analysis revealed that these species were linked to Tolypocladium and several species of Verticillium [Citation3]. The species of Cordyceps sensu lato that parasitizes ectomycorrhizal Elaphomyces (2 forma and 18 species), beetle larvae (Cordyceps subsessilis), and cicada nymphs (Cordyceps inegonsis, C. paradoxa, and C. toriharamontana) was also reassigned to Elaphocordyceps [Citation3]. Moreover, Chaunopycnis was reclassified to accommodate Ch. alba, a conidiogenically similar species to Tolypocladium [Citation6]. Following the “One fungus, One name” principle, the genera Elaphocordyceps and Chaunopycnis were merged under the genus Tolypocladium due to the latter being the oldest, most widely recognized, and having therapeutic value [Citation2–4,Citation6]. Most Tolypocladium species, except for a small number of entomopathogens, are mycoparasites of the truffle-like ectomycorrhizal ascomycete Elaphomyces (Elaphomycetaceae, Eurotiales). Previously identified as Elaphocordyceps spp., those mycoparasites have recently been given the name Tolypocladium [Citation4]. Identification of Tolypocladium species solely based on morphological traits has many limitations, leading to misidentification of some species. The asexual morphological structure of all identified Tolypocladium species has not yet been reported. Several species have been reidentified and moved into or out of the genus because of the development of molecular biology techniques, which is based on the combination of conventional morphological traits and molecular markers [Citation3,Citation4,Citation7].
This study aimed to discover unidentified fungal species from Korea. Morphological and cultural characteristics as well as molecular phylogenetic analysis were used to confirm the novelty of the obtained strains.
2. Materials and methods
2.1. Collection of soil samples and fungal isolation
The soil samples were obtained in 2022 from two locations in Korea: Chiltansan, Miryang-si, Gyeongsangnam-do (35°28′41.9″N, 128°49′33.2″E) and Sobaeksan National Park, Yeongju-si, Gyeongsangbuk-do (36°56′44.9″N, 128°27′43.9″E). Then, the isolates were processed using the standard serial dilution method [Citation8]. A vortex was used to completely mix 1 g of soil with 9 mL of double-distilled water for 1–3 min. The soil suspension was serially diluted in double-distilled water, with 1000 µL of each sample being diluted to 10−1, 10−2, 10−3, and 10−4. Potato dextrose agar (PDA; Difco, Detroit, MI, USA) plates were pipetted with 100 μL of 10−3 and 10−4 suspensions, and then the plates were incubated in an incubator at 25 °C. Fungi grew on PDA plates after 3–4 days of incubation. Selected fungal colonies were subcultured onto new PDA media and incubated under the same previous conditions for subsequent examination.
2.2. Cultural and morphological characterization
For the study of cultural characteristics and morphological characteristics, PDA, malt extract agar (MEA; Difco, Detroit, MI, USA), and cornmeal agar (CMA; Difco, Detroit, MI, USA) were used [Citation2,Citation9–11]. The KNUF-22-14A and KNUF-22-15A strains were incubated for 14 days at 25 °C. After measuring the diameter of the colonies and noting their characteristics, mycological characteristics were observed by examining the fungal structures using a light microscope (BX-50; Olympus, Tokyo, Japan).
2.3. Genomic DNA extraction, PCR amplification, and sequencing
For genomic DNA extraction, a HiGene Genomic DNA prep kit (BIOFACT, Daejeon, Korea) was used in accordance with the manufacturer’s instructions, and the obtained sample was kept at −20 °C until use. KNUF-22-14A and KNUF-22-15A were amplified via polymerase chain reaction (PCR) with the dataset of internal transcribed spacer (ITS) regions, large subunit of 28S rRNA (LSU), and small subunit of 18S rRNA (SSU). The primer pairs ITS1F/ITS4, LROR/LR5, and NS1/NS8 were employed for ITS, LSU, and SSU, respectively [Citation12–15]. The thermal conditions for PCR amplification were set as previously described [Citation16]. EXOSAP-IT (Thermo Fisher Scientific, Waltham, MA, USA) was used to purify the amplified PCR products and sequenced by Solgent Co. Ltd. (Daejeon, Korea). Using the software SeqMan Lasergene (DNAStar Inc., Madison, WI, USA), the sequencing data collected for this investigation were adjusted. The resulting sequences for the ITS regions, LSU, and SSU were deposited in National Center for Biotechnology Information (NCBI) GenBank under the following accession numbers: KNUF-22-14A (LC731698, LC731699, and LC731700) and KNUF-22-15A (LC731703, LC731704, and LC731705), respectively.
2.4. Molecular phylogenetic analyses
Using the basic local alignment search tool (BLAST), three molecular markers (ITS, LSU, and SSU) were used and compared the reference sequences with KNUF-22-14A and KNUF-22-15A from the GenBank database of the NCBI (). Evolutionary distance matrices were generated based on the Kimura’s neighbor-joining algorithm model [Citation17]. To determine the precise taxonomic position of each strain, neighbor-joining [Citation18], maximum likelihood [Citation19], and maximum parsimony [Citation16] trees were constructed. The MEGA 7.0 software program was used for the phylogenetic analyses, and bootstrap values based on 1000 replications were used [Citation20].
Table 1. GenBank accession numbers of fungal strains used in this study for phylogenetic analysis.
3. Results
3.1. Taxonomic analysis of Tolypocladium globosum sp. nov.
Molecular phylogenetic analyses revealed that the strains KNUF-22-14A and KNUF-22-15A were similar as well as clustered together. Thus, only cultural, and morphological characteristics for the taxonomic descriptions and microphotographs as type strain KNUF-22-14A are described in this paper ( and ).
Figure 1. Cultural and morphological characteristics of Tolypocladium globosum (KNUF-22-14AT). Colonies on potato dextrose agar (A); malt extract agar (B); and cornmeal agar (C) following 14 days of incubation at 25 °C, respectively. Phialides (D–H); conidia (I,J). Scale bars: D = 20 µm, E–J = 10 µm.
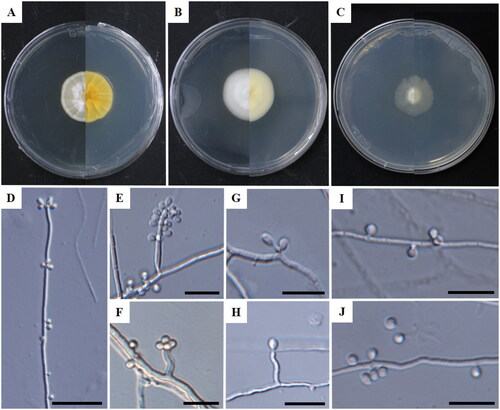
Table 2. Morphological comparison of Tolypocladium globosum (KNUF-22-14AT) with the closest species of Tolypocladium.
Tolypocladium globosum J. J. Ryu, S.Y. Lee, and H.Y. Jung, sp. nov. ()
MycoBank: MB 845983
Etymology: The epithet “globosum” specifically refers to conidia with a globose morphology.
Typus: KNUF-22-14A and KNUF-22-15A strains were isolated from soil in Sobaeksan National Park, Yeongju-si, Gyeongsangbuk-do (36°56′44.9″N, 128°27′43.9″E) and Chiltansan, Miryang-si, Gyeongsangnam-do (35°28′41.9″N, 128°49′33.2″E) in Korea, respectively. The stock culture (NIBRFGC000509972) was deposited in the National Institute of Biological Resources (NIBR), as a metabolically inactive culture.
Ecology and distribution: Several species from this genus were isolated from the soil, sapwood, and decaying wood. In addition, several species were discovered from Elaphomyces sp., also referred to as deer truffles. Moreover, numerous individuals were isolated from Cicada nymphs and Scarabaeidae larvae (e.g., Auritibicen bihamatus, Graptopsaltria nigrofuscata, and Platypleura kaempferi). The novel species proposed in this study, Tolypocladium globosum, was found in soil that was collected from mountains in Korea.
Cultural characteristics: On PDA, the colonies were flat, white to light beige, round, wrinkled, reverse colonies were yellow, and they had grown to a diameter of 29–30.5 mm (). On MEA, the surface was white, floccose, whereas the reverse surface was pale yellow to light beige, with colonies growing to 30–31.5 mm in diameter (). On CMA, the colonies grew very slowly, the surface was floccose and mycelium white, whereas the reverse surface was light yellow to pale yellow, with colonies measuring 19–21 mm in diameter (). All media were incubated for 14 days at 25 °C.
Morphological characteristics: The colonies grown on PDA were used to study the morphological structures. Conidiomata were absent, branched hyphae, hyaline, smooth-walled, 1.2–2.4 µm wide. Phialides were 0.6–2.4(–9.3) × 0.8–1.4 µm long, swollen, curved or erect, and short phialides were commonly produced attached to conidiophores (). Conidiophores were micronematous, erect, and up to 314 µm in length. Conidia were solitary or verticils of 2 to 6, hyaline, globose to subglobose, and with a diameter of 1.2–3.0 × 1.2–3.0 µm (x̅ = 2.1 × 2.0 µm, n = 100), L/W ratio of 1.0 ().
Note: The strain KNUF-22-14A was morphologically compared with phylogenetically related strains: Tolypocladium album, T. amazonense, T. endophyticum, T. pustulatum, and T. tropicale (). Morphological differences were observed between KNUF-22-14A and related species. The cultural characteristics of KNUF-22-14A on CMA were flat, floccose, irregular margin, hyaline in reverse, and colonies reaching 22–24 mm in diameter after 14 days at 25 °C, while the phylogenetically closest strain T. pustulatum (MRL GB6597) exhibited appressed, margin even, hyaline, powdery or farinaceous, conidial pustules light pink, dull grey, hyaline in reverse after one month, reaching 21–22 mm in diameter after 14 days at 25 °C on CMA. T. album (CCF 3185) displayed white, floccose, with a yellow tint in reverse, growing rather slowly, reaching 31–35 mm on CMA with a diameter after 14 days at 25 °C [Citation9,Citation10]. Other related strains T. endophyticum, T. amazonense, and T. tropicale showed differences in phialides morphology, forming a trichodermoid branching pattern of phialides [Citation7]. KNUF-22-14A [0.6–2.4(–9.3) × 0.8–1.4 µm] had short or curved phialides attached to conidiophores that are shorter than those of T. album (3.5–10.0 × 1.0–1.5 µm), T. amazonense (4.6 ± 1.2 × 1.5 ± 0.3 µm), T. endophyticum (4.1 ± 0.9 × 1.6 ± 0.2 µm), T. pustulatum (4.0–10.0 × 2.0–4.0 µm), and T. tropicale (4.6 ± 1.2 × 1.5 ± 0.3 µm) (). In addition, KNUF-22-14A (1.2–3.0 × 1.2–3.0 µm) can be distinguished from T. album (1.6–1.9 µm), T. amazonense (2.1–2.2 µm), T. endophyticum (1.9–2.0 µm), T. tropicale (2.0–2.1 µm), and T. pustulatum (2.0–3.0 × 1.5–2.5 µm) based on their larger conidia. Moreover, T. pustulatum produced conidia accumulating in dry chains, but KNUF-22-14A did not produce chains of conidia. Thus, the morphological characteristics of KNUF-22-14A were distinct from those of previously reported Tolypocladium species.
3.2. Molecular phylogeny of the strain KNUF-22-14A
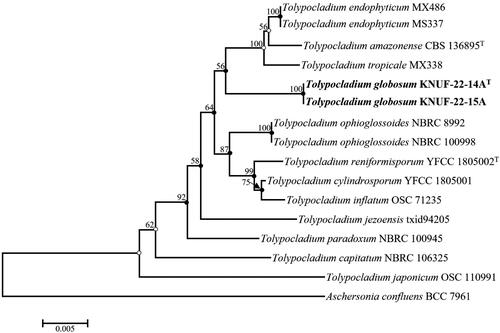
The novel species in this genus were investigated using BLAST tool from NCBI, and the molecular markers ITS regions, SSU, and LSU genes were examined. According to the sequencing results, the lengths of the sequences for KNUF-22-14A and KNUF-22-15A were as follows: SSU (1659, 1650 bp), LSU (831, 857 bp), ITS (595, 596 bp), TEF1 (871, 895 bp), and RPB2 (1083, 1106 bp), respectively. The BLAST search results of KNUF-22-14A and KNUF-22-15A in ITS regions showed 95% similarity with T. endophyticum (MX 485) and T. inflatum (PANM200T9ZM1D). In SSU, T. cylindrosporum (NBRC 100548) and T. inflatum (NBRC 31671) showed maximal similarity of 99.7% and 99.8%, respectively. Similarly, based on the LSU gene sequencing results of KNUF-22-14A and KNUF-22-15A, highest similarity (99.0% and 99.1%) was displayed with Tolypocladium paradoxum (NBRC 100945). A phylogenetic tree based on ITS regions was made to shed more resolution on the phylogenetic positions of the newly obtained strains along with additional taxa, since not many Tolypocladium species contain gene sequences that are not listed in GenBank ( and ). The strains KNUF-22-14A and KNUF-22-15A developed individual lineages with the Tolypocladium species T. album, T. amazonense, T. endophyticum, T. pustulatum, and T. tropicale. In addition, a phylogenetic tree was created based on the neighbor-joining method using 16 closely related fungal taxa from the Tolypocladium genus to perform multi-locus molecular markers (ITS, SSU, and LSU) of the strains KNUF-22-14A and KNUF-22-15A. This tree also clearly demonstrated that the strains KNUF-22-14A and KNUF-22-15A occupied a distinct position from other Tolypocladium species and phylogenetically closest to T. amazonense, T. endophyticum, and T. tropicale (). Maximum likelihood and maximum parsimony trees were also constructed to determine the exact taxonomic position of the strains; and filled circles in the neighbor-joining phylogenetic tree indicated the nodes, whereas open circles indicated the corresponding nodes with the maximum likelihood or maximum parsimony algorithm (). From the two phylogenetic trees, it is clearly shown that the strains KNUF-22-14A and KNUF-22-15A have distinct positions compared with previously identified Tolypocladium species.
Figure 2. Neighbor-joining phylogenetic tree based on the concatenated sequences of ITS sequences showing the phylogenetic position of strains KNUF-22-14AT and KNUF-22-15A among Tolypocladium species. Bootstrap values based on 1000 replicates are shown next to the branches. Aschersonia confluens BCC 7961 comprised the outgroup. Bootstrap values greater than 50% (based on 1000 replications) are shown at the branching points. The isolated strains are shown in bold. Bar = 0.01 substitutions per nucleotide position.
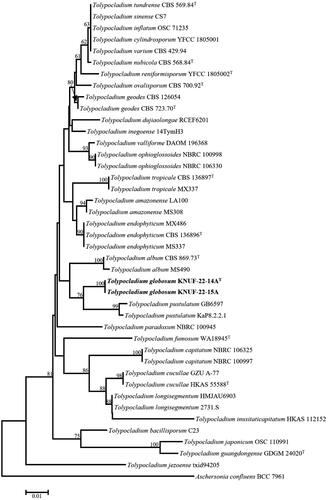
Figure 3. Neighbor-joining phylogenetic tree based on the combined molecular markers of the internal transcribed spacer (ITS) regions, small subunit (SSU), and large subunit (LSU) showing the phylogenetic position of strains KNUF-22-14AT and KNUF-22-15A among Tolypocladium species. Aschersonia confluens BCC 7961 comprised the outgroup. Bootstrap values greater than 50% (based on 1000 replications) are shown at the branching points. The neighbor-joining tree, maximum likelihood, and maximum parsimony trees indicated with filled nodes and open circles were made using maximum likelihood or parsimony. The isolated strains are shown in bold. Bar = 0.005 substitutions per nucleotide position.
4. Discussion
In the present study, two strains KNUF-22-14A and KNUF-22-15A were isolated in 2022 from soil samples in Yeongju and Miryang, South Korea. The strains exhibited morphological differences from each of the previously identified closely related species, as supported by previous descriptions of the latter ().
Species with known sexual stages have been used mainly in the studies of the ecology of the Tolypocladium clade. Most species of Tolypocladium are parasites of the truffle-like fungal genus Elaphomyces and to a lesser extent of soil-inhabiting Cicada nymphs (Cicadidae, Hemiptera) and wood-inhabiting beetle larvae (Coleoptera) [Citation3]. Members of this clade and the Ophiocordycipitaceae are well recognized for their symbiosis with hosts, which is often host-specific. Elaphomyces species are the only fungi documented as hosts for Tolypocladium to date [Citation21]. The same species can be found playing different ecological roles that are frequently connected to various substrates by asexual and sexual states. They are capable of producing metabolites with significant medical and biocontrol benefits, such as tolypin, tolypoalbin, tolyprolinol, cyclosporin A, and cyclosporin D hydroperoxide [Citation22–26]. The type species of the genus, T. inflatum, can produce cyclosporin A. This is one of the most often prescribed immunosuppressants for people with autoimmune conditions, including organ transplant recipients and those with AIDS, due to its superior T cell specificity and low levels of myelotoxicity [Citation27]. In addition, T. cylindrosporum has the potential to be used as a biological control agent as it kills mosquito eggs, larvae, and adults [Citation28]. Numerous secondary metabolites, such as cyclosporin, efrapeptins, ophiocordin, and ophiosetin, are produced by several Tolypocladium species [Citation29]. They have been broadly applied in both biocontrol and biopharmaceuticals [Citation29]. An unidentified Tolypocladium sp. was discovered in the roots of plants (e.g., Isachne globosa, Scirpus karuisawensis, Utricularia racemosa, and Eriocaulon decemflorum) from Jangdo Island in Korea, along with various other unidentified fungi. Other Tolypocladium species, including T. inegoense, T. ophioglossoides, and T. paradoxum, have been isolated in Korea [Citation30]. In addition, Chaunopycnis alba (also known as T. album) was isolated from soil for the first time in Korea [Citation31].
In conclusion, the strains obtained in this study were recognized as novel species based on cultural, morphological, and phylogenetic analyses. These results revealed that the strains differed significantly from the previously described Tolypocladium species. The etiology of these two species as well as their pathogenicity, ecological significance in Korean soils, environmental conditions, and relationships with agricultural production need to be studied further.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Sung GH, Poinar GO, Spatafora JW. The oldest fossil evidence of animal parasitism by fungi supports a cretaceous diversification of fungal-arthropod symbioses. Mol Phylogenet Evol. 2008;49(2):495–502.
- Gams W. Tolypocladium, eine Hyphomycetengattung mit geschwollenen Phialiden. Persoonia. 1971;6:185–191.
- Sung GH, Hywel-Jones NL, Sung JM, et al. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud Mycol. 2007;57:5–59.
- Quandt CA, Kepler RM, Gams W, et al. Phylogenetic-based nomenclatural proposals for Ophiocordycipitaceae (Hypocreales) with new combinations in Tolypocladium. IMA Fungus. 2014;5(1):121–134.
- Hodge KT, Krasnoff SB, Humber RA. Tolypocladium inflatum is the anamorph of Cordyceps subsessilis. Mycologia. 1996;88(5):715–719.
- Gams W. Chaunopycnis alba, gen. et sp. nov., a soil fungus intermediate between Moniliales and Sphaeropsidales. Persoonia. 1980;11:75–79.
- Gazis R, Skaltsas D, Chaverri P. Novel endophytic lineages of Tolypocladium provide new insights into the ecology and evolution of Cordyceps-like fungi. Mycologia. 2014;106(6):1090–1105.
- Park S, Ten L, Lee SY, et al. New recorded species in three genera of the Sordariomycetes in Korea. Mycobiology. 2017;45(2):64–72.
- Kubátová A, Černý M, Nováková A. New records of micromycetes from the Czech Republic. IV. Acrodontium salmoneum, Chaunopycnis alba and Cylindrocarpostylus gregarius, and notes on Dactylaria lanosa and Trichoderma saturnisporum. Czech Mycol. 2002;53(3):237–255.
- Bills GF, Polishook JD, Goetz MA, et al. Chaunopycnis pustulata sp. nov., a new clavicipitalean anamorph producing metabolites that modulate potassium ion channels. Mycol Progress. 2002;1(1):3–17.
- Wang Y, Liu YF, Tang DX, et al. Tolypocladium reniformisporum sp. nov. and Tolypocladium cylindrosporum (Ophiocordycipitaceae, Hypocreales) co-occurring on Ophiocordyceps sinensis. Mycol Progress. 2022;21(1):199–214.
- Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172(8):4238–4246.
- White TJ, Bruns TD, Lee SB, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego (CA): Academic Press; 1990. p. 315–322.
- Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2(2):113–118.
- Wijayawardene NN, Dissanayake LS, Li QR, et al. Yunnan– Guizhou Plateau: a mycological hotspot. Phytotaxa. 2021;523(1):1–31.
- Fitch WM. Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool. 1971;20(4):406–416.
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111–120.
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425.
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17(6):368–376.
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874.
- Spatafora JW, Sung GH, Sung JM, et al. Phylogenetic evidence for an animal pathogen origin of ergot and the grass endophytes. Mol Ecol. 2007;16(8):1701–1711.
- Weiser J, Matha V. Tolypin, a new insecticidal metabolite of fungi of the genus Tolypocladium. J Invertebr Pathol. 1988;51(1):94–96.
- Sedmera P, Havlicek V, Jegorov A, et al. Cyclosporin D hydroperoxide, a new metabolite of Tolypocladium terricola. Tetrahedron Lett. 1995;36:6953–6956.
- Bushley KE, Raja R, Jaiswal P, et al. The genome of Tolypocladium inflatum: evolution, organization, and expression of the cyclosporin biosynthetic gene cluster. PLoS Genet. 2013;9(6):e1003496.
- Fukuda T, Sudoh Y, Tsuchiya Y, et al. Tolypoalbin, a new tetramic acid from Tolypocladium album TAMA 479. J Antibiot. 2015;68(6):399–402.
- Fukasawa W, Mori N, Iwatsuki M, et al. Tolyprolinol, a new dipeptide from Tolypocladium sp. FKI-7981. J Antibiot. 2018;71:682–684.
- Survase SA, Kagliwal LD, Annapure US, et al. Cyclosporin A - a review on fermentative production, downstream processing and pharmacological applications. Biotechnol Adv. 2011;29(4):418–435.
- Laupacis A, Keown PA, Ulan RA, et al. Cyclosporin A: a powerful immunosuppressant. Can Med Assoc J. 1982;126:1041–1046.
- Wang JC, Zhang ZZ, Li CL, et al. Research progress of Tolypocladium in Ophiocordycipitaceae. J Fungal Res. 2019;8:1–0.
- Park JM, Hong JW, Lee W, et al. Fungal clusters and their uniqueness in geographically segregated wetlands: a step forward to marsh conservation for a wealth of future fungal resources. Mycobiology. 2020;48(5):351–363.
- Das K, Lee SY, Jung HY. A new report on three species of sordariomycetes class isolated from soil in Korea. Kor J Mycol. 2017;46:134–144.
