Abstract
During disease surveys of Angelica acutiloba plants in Korea, leaf spot symptoms were observed in a field in Andong in July 2019, and stem rot symptoms in vinyl greenhouses in Yangpyeong in April 2020. Incidence of leaf spot and stem rot of the plants ranged from 10 to 20% and 5 to 30%, respectively. Morphological and cultural characteristics of fungal isolates from the leaf spot and stem rot symptoms fitted into those of the genus Phoma. Molecular phylogenetic analyses of two single-spore isolates from the symptoms using concatenated sequences of LSU, ITS, TUB2, and RPB2 genes authenticated an independent cluster from other Didymella (anamorph: Phoma) species. Moreover, the isolates showed different morphological and cultural characteristics in comparison to closely related Didymella species. These discoveries confirmed the novelty of the isolates. Pathogenicity of the novel Didymella species isolates was substantiated on leaves and stems of A. acutiloba through artificial inoculation. Thus, this study reveals that Didymella acutilobae sp. nov. causes leaf spot and stem rot in Angelica acutiloba.
1. Introduction
Angelica acutiloba (Siebold & Zucc.) Kitag. is a perennial plant belonging to the family Apiaceae. This plant is native to Japan and has been introduced to Korea and Manchuria [Citation1]. It has been traditionally utilized as a medicinal herb with various pharmacological effects in Japan [Citation2] and is cultivated as a vegetable in Korea.
We observed leaf spot symptoms with a yellow halo on A. acutiloba plants in a field in Andong, Gyeongbuk Province, Korea, during a disease survey in July 2019. We also observed stem rot symptoms on the plants in vinyl greenhouses in Yangpyeong, Gyeonggi Province, Korea, during a disease survey in April 2020. We obtained fungal isolates from the leaf spot and stem rot symptoms and examined morphological characteristics of the isolates. The morphological and cultural characteristics of the isolates fitted into those of the genus Phoma [Citation3]. The isolates were very similar to each other in terms of morphological characteristics.
Morphological characteristics, host relationships, and cultural traits were investigated in the beginning stage for identification of Phoma spp. [Citation3,Citation4]. In addition, the genus Didymella was revealed as a teleomorph of the genus Phoma based on the phylogenetic studies [Citation4,Citation5]. Many reported Phoma spp. have been re-identified into new genus or species gradually based on molecular phylogenetic studies [Citation6–9].
It has been reported that Didymella sp. causes leaf spot of A. acutiloba in Japan [Citation10]. In Korea, stem blight of A. acutiloba caused by Phoma sp. is recorded [Citation11]. However, no significant study has not been proceeded to identify a causal agent of leaf spot and stem rot in the plant. This study was conducted to identify Phoma sp. isolates from leaf spot and stem rot symptoms of A. acutiloba in Korea using a multi-locus phylogenetic analysis with investigations of morphological and cultural characteristics. In addition, pathogenicity test of the isolates was carried out to reveal the causal agent of the leaf spot and stem rot in A. acutiloba.
2. Materials and methods
2.1. Disease survey and fungal isolation
We surveyed occurrence of diseases on A. acutiloba plants grown in a field in Andong, Gyeongbuk Province, Korea in July 2019 and three vinyl greenhouses in Yangpyeong, Gyeonggi Province, Korea in April 2020. One hundred leaves of the plants were observed for investigation of leaf spot occurrence in the field, and 100 plants for investigation of stem rot occurrence in the vinyl greenhouses. The investigation for occurrence of leaf spot and stem rot was conducted at three sites in the field or vinyl greenhouse. Lesion pieces (5–6 mm2) were cut from the leaves and stems of the diseased plants and surface-sterilized with 1% sodium hypochlorite solution for one minute. The sterilized pieces were placed on 2% water agar (WA) and incubated at 25 °C for 3–4 days. Then, growing mycelia from the pieces were transferred onto potato dextrose agar (PDA). The isolates were transferred onto oatmeal agar (OA), and single-spore isolates were obtained from 2-week-old OA cultures. One isolate from the leaf and stem, respectively, was selected among the single-spore isolates for identification and pathogenicity tests. A representative isolate was deposited in the Korean Agricultural Culture Collection (KACC), Wanju, Korea.
2.2. Investigation of cultural and morphological characteristics
Malt extracted agar (MEA), OA, and PDA were used to investigate cultural characteristics of the isolates (ANAC-1901 and ANAC-2001). The isolates were incubated on the media at 22 °C in the dark for seven days. Subsequently, another week of incubation was continued at 22 °C under alternating cycles of 13/11 h of near ultraviolet light and dark [Citation6,Citation7]. NaOH spot test [Citation6] was performed on 1-week-old culture on MEA. Twenty pycnidia and 30 conidia from 2-week-old cultures on OA were observed to investigate their morphological characteristics under the light microscope (Nikon Eclipse Ci-L, Tokyo, Japan).
2.3. DNA extraction, PCR, and sequencing
Genomic DNA of the isolates was extracted using the protocol in a previous study [Citation12], with slight modifications. Polymerase chain reaction (PCR) experiments were conducted to investigate partial large subunit nuclear ribosomal DNA (LSU), internal transcribed spacer regions 1 and 2 including 5.8S nrDNA (ITS), β-tubulin (TUB2), and RNA polymerase II second largest subunit (RPB2) gene regions using LR0R [Citation13] and LR7 [Citation14] for LSU, V9G [Citation15] and ITS4 [Citation16] for ITS, Btub2Fd and Btub4Rd [Citation17] for TUB2, and RPB2-5f2 [Citation18] and fRPB2-7cR [Citation19] for RPB2. Conditions of PCR amplification for all the genes were followed as in the previous studies [Citation7]. Takara Ex Taq (Takara Bio Inc., Kusatsu, Japan) was used to prepare PCR products of the two isolates following the manufacturer’s instruction. Purification of the PCR products was conducted using the universal DNA purification kit (Tiangen, Beijing, China), according to the manufacturer’s instruction. The PCR products were sequenced using the same primers at Bionics Co., Ltd. (Seoul, Korea). The sequences were improved manually when necessary by SeqMan II (DNASTAR Inc., Madison, WI). The sequence data were deposited in GenBank.
2.4. Alignment and molecular phylogenetic analysis
The sequences of the isolates obtained from A. acutiloba and the pertinent sequences of Didymella spp. from the previous studies [Citation7–9,Citation20] () were aligned by MUSCLE [Citation21]. Coniothyrium palmarum (CBS 400.71) was served as an outgroup taxon. The multiple sequence alignments were conducted and improved when necessary using MEGA version 7 software [Citation22]. The partition-homogeneity test for the sequences was carried out using PAUP version 4.0 software [Citation23]. Then, neighbor-joining (NJ) analysis with the maximum composite likelihood model was performed with 1000 bootstrap (BS) replicates using MEGA version 7 software [Citation22]. Bootstrap values equal to or greater than 50% were indicated at nodes. The evolutionary model for each gene was investigated using MrModeltest version 2.2 software [Citation24]. The Bayesian analysis of the concatenated alignments was conducted by MrBayes version 3.2.4 software [Citation25] based on the results of the model test. The calculation continued until the average standard deviation of split frequencies reached a value of equal to or less than 0.01. Generated trees were taken 25% burn-in process to calculate posterior probabilities (PPs). The probabilities equal to or greater than 0.9 were displayed at the nodes. The tree was visualized using FigTree version 1.4.4 software [Citation26].
Table 1. Isolates of Didymella spp. and Coniothyrium palmarum used for molecular phylogenetic analyses in this study.
2.5. Pathogenicity test
One isolate from the leaf and stem of A. acutiloba, respectively, was used to verify its pathogenicity on leaves and stems of the host plant. Conidial suspension of each isolate harvested from 2-week-old cultures on OA was filtered through two layers of Miracloth (Sigma-Aldrich, St. Louis, MO) and suspended in sterile distilled water. A. acutiloba plants were grown in plastic pots (height: 14 cm; upper diameter: 15 cm; lower diameter: 10 cm) with commercial media in a vinyl greenhouse. A 20 ml conidial suspension (1–2 × 106 conidia/ml) of each isolate was sprayed onto the leaves of 4-month-old A. acutiloba plants after shooting for inoculation test to leaves. A 10 ml conidial suspension (1–2 × 107 conidia/ml) of each isolate was sprayed onto the stems at the soil surface level of the A. acutiloba plants for inoculation test to stems. The inoculated plants were placed in plastic boxes (63 × 44 × 47 cm) under 100% relative humidity at room temperature (24–26 °C). Control plants were sprayed with the same quantity of sterile distilled water and placed under the same conditions as the inoculated plants. After five days, the inoculated plants were taken out from the boxes and kept indoors. The pathogenicity of the isolates was assessed based on the induced symptoms 10–12 days after inoculation. The inoculation test was performed in triplicate.
3. Results
3.1. Molecular phylogeny
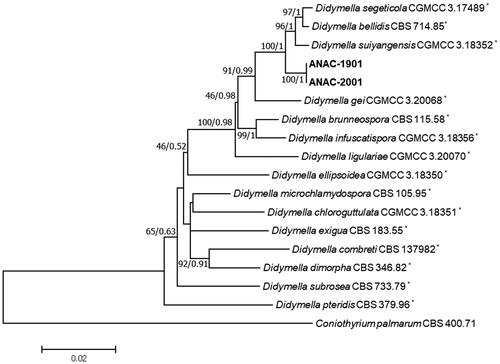
According to the model test, the best fitting model for sequences of LSU, ITS, TUB2, and RPB2 were detected as HKY + I, HKY + I + G, GTR + I, and GTR + G, respectively. The partition-homogeneity test (p value = 0.94) confirmed that the phylogenetic trees of each gene have a common underlying structure, which enable us to concatenate the alignments of each gene. The concatenated sequences of the isolates and relevant Didymella spp. contained a total 2130 characters (794, 453, 285, and 599 characters of LSU, ITS, TUB2, and RPB2, respectively). Two analyses indicated no significant differences in tree topology (data not shown). Hence, only the NJ tree with BS values was shown with PPs at nodes (BS/PP). The representative isolates, ANAC-1901 and ANAC-2001 obtained from leaf spot and stem rot symptoms, respectively, were determined as members of the genus Didymella (). The isolates were positioned in a group and verified as the same species because of no difference in their sequences. In addition, the isolates were not categorized as any species within the genus. Furthermore, a single clade consisting of the isolates was constructed with a high BS value and PP that was clearly separated from other closely related species such as Didymella bellidis (Neerg.) Qian Chen & L. Cai (anamorph: Phoma bellidis Neerg.) [Citation7,Citation27], Didymella segeticola (Q. Chen) Q. Chen, Crous & L. Cai (anamorph: Phoma segeticola) [Citation8,Citation28], and Didymella suiyangensis Qian Chen, Crous & L. Cai [Citation8]. GenBank accession numbers of the isolates, ANAC-1901 and ANAC-2001 are OQ749982–OQ749983, OQ749980–OQ749981, OQ744070–OQ744071, and OQ744072–OQ744073 for LSU, ITS, TUB2, and RPB2, respectively.
Figure 1. A phylogenetic tree generated from the neighbor-joining analysis with maximum composite likelihood model based on a concatenated alignment of partial large subunit nuclear ribosomal DNA, internal transcribed spacer regions 1 and 2 including 5.8S nrDNA, β-tubulin, and RNA polymerase II second largest subunit sequences of two isolates (ANAC-1901 and ANAC-2001) of Didymella acutilobae sp. nov. and relevant Didymella spp. Bootstrap values (BS) and posterior probabilities (PP) are given at nodes (BS/PP). The bar represents the number of nucleotide substitutions per site. The phylogenetic tree was rooted to Coniothyrium palmarum (CBS 400.71). *The reference strains.
3.2. Taxonomy
Didymella acutilobae G.B. Lee and W.G. Kim, sp. nov. ()
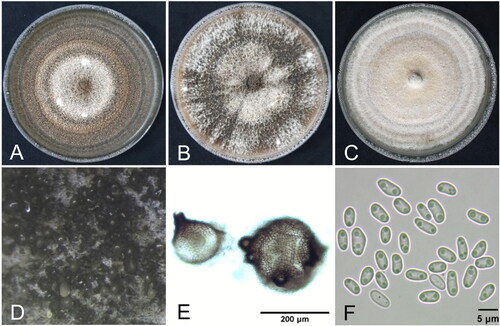
Figure 2. Cultural and morphological features of Didymella acutilobae sp. nov. (A) Two-week-old colonies on malt extract agar; (B) oatmeal agar; (C) potato dextrose agar. (D, E) Pycnidia produced in oatmeal agar. (F) Conidia.
MycoBank no.: MB 849458.
Etymology: Named derived from specific epithet of the host plant, Angelica acutiloba.
Holotype: Isolated from stem of A. acutiloba, Yangpyeong, Gyeonggi Province, Korea (37° 27′ 51″ N and 127° 33′ 16.999″ E), April 2020, W.G. Kim, ex-holotype culture (KACC 410302).
Cultural and morphological characteristics: Diameter of 1-week-old cultures on MEA, OA, and PDA was 54–61 mm (av. 58 mm), 53–54 mm (av. 54 mm), and 53–55 mm (av. 53 mm), respectively. The culture on MEA showed brown to black colony with white mycelium and light black concentric rings (). The culture on OA showed brown to dark olivaceous colony with white mycelium (). The culture on PDA showed white to light brown colony with white mycelium and whitish brown concentric rings (). NaOH spot test on MEA was negative.
Teleomorph was not observed in the cultures. Pycnidia usually half submerged in the agar or on the surface (), 70–240 μm in diameter, solitary or confluent, globose, brown to black, with 1–5 ostioles, non-papillate or papillate (). Conidia 2.9–6.5 × 1.6–3.0 μm (av. 4.7 × 2.3 μm), ellipsoidal or slightly curved, aseptate with usually two bipolar guttules (). Conidial matrix white. Chlamydospores absent. Summarized descriptions on the cultural and morphological characteristics of D. acutilobae and closely related Didymella spp. are shown in . D. acutilobae was somewhat dissimilar to the closely related Didymella spp. in the cultural and morphological characteristics.
Table 2. Summarized descriptions on the cultural and morphological characteristics of Didymella acutilobae and closely related Didymella species.
3.3. Disease incidence and pathogenicity
During the disease surveys in Korea, leaf spot symptoms were found on A. acutiloba plants in the investigated field in Andong, Gyeongbuk Province in July 2019, and stem rot symptoms in the investigated vinyl greenhouses in Yangpyeong, Gyeonggi Province in April 2020. The leaf spot symptoms were circular to elliptical, brown to dark brown with a yellow halo, and 2–5 mm in diameter in the early stage (). In the late stage, they enlarged to irregular shape of 10–20 mm in diameter. The stem rot symptoms showed wilt and blight due to rot of the lower part of the stem (). Incidence of leaf spot and stem rot of the plants during the disease surveys ranged from 10 to 20% and 5 to 30%, respectively.
Figure 3. Leaf spot and stem rot symptoms of Angelica acutiloba plants. (A, B) Symptoms on the leaves and stems observed in the investigated field and vinyl greenhouse, respectively. Induced symptoms on the leaf (C) and the stem (D) by artificial inoculation with the isolates of Didymella acutilobae sp. nov. in pathogenicity tests. The white arrow (D) indicates a stem rot lesion formed on the stem. (E, F) Non-inoculated control plants.

The isolates (ANAC-1901 and ANAC-2001) from leaf and stem rot lesions of A. acutiloba induced leaf spot and stem rot symptoms in the inoculated plants of A. acutiloba (), but no symptoms were observed on the leaves and stems of the control plants (). The induced symptoms were similar to those observed on the plants of A. acutiloba during the disease surveys. Re-isolation of the isolates from the induced leaf spot and stem rot lesions was confirmed based on morphological characteristics.
4. Discussion
According to the phylogenetic analyses, two isolates of D. acutilobae were located in the genus Didymella with positioning in as an independent group from other closely related species such as D. bellidis, D. segeticola, and D. suiyangensis. Compared to morphology of D. bellidis, D. segiticola, and D. suiyangensis [Citation7,Citation8,Citation27,Citation28], D. acutilobae produced much smaller conidia than those of D. segeticola and D. suiyangensis. D. acutilobae developed much bigger pycnidia than D. segeticola and showed more ostioles of pycnidia than D. segeticola and D. suiyangensis. The growth rates of D. acutilobae on MEA, OA, and PDA were generally much slower than those of D. bellidis, D. segeticola, and D. suiyangensis. In NaOH spot test, only D. acutilobae and D. segeticola exhibited negative reaction among closely related Didymella spp. Based on the phylogenetic tree analyses and investigations of the morphological characteristics, it is suggested that the isolates signify a novel species in the genus Didymella. Hence, we propose Didymella acutilobae sp. nov. as a fungal pathogen causing leaf spot and stem rot in Angelica acutiloba.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Plants of the World Online. Angelica acutiloba (Siebold & Zucc.) Kitag. [Internet]. Kew: Royal Botanic Gardens; 2022 [cited 2022 Aug 8]. Available from: https://powo.science.kew.org/taxon/urn:lsid:ipni.org:names:837548-1
- Takeda Chemical Industries. List of plants. Ichijoji: Kyoto Herbal Garden, Pharmacognostic Research Lab., Central Research Division, Takeda Chem. Industries, Ltd.; 1978.
- Boerema GH, De Gruyter J, Noordeloos ME, et al. Phoma identification manual. Differentiation of specific and infra-specific taxa in culture. Oxfordshire: CABI Publishing; 2004.
- Aveskamp MM, De Gruyter J, Crous PW. Biology and recent developments in the systematics of Phoma, a complex genus of major quarantine significance. Fungal Divers. 2008;31:1–18.
- De Gruyter J, Aveskamp MM, Woudenberg JHC, et al. Molecular phylogeny of Phoma and allied anamorph genera: towards a reclassification of the Phoma complex. Mycol Res. 2009;113:508–519. doi: 10.1016/j.mycres.2009.01.002.
- Aveskamp MM, De Gruyter J, Woudenberg JHC, et al. Highlights of the Didymellaceae: a polyphasic approach to characterise Phoma and related pleosporalean genera. Stud Mycol. 2010;65:1–60. doi: 10.3114/sim.2010.65.01.
- Chen Q, Jiang JR, Zhang GZ, et al. Resolving the Phoma enigma. Stud Mycol. 2015;82(1):137–217. doi: 10.1016/j.simyco.2015.10.003.
- Chen Q, Hou LW, Duan WJ, et al. Didymellaceae revisited. Stud Mycol. 2017;87(1):105–159. doi: 10.1016/j.simyco.2017.06.002.
- Hou LW, Groenewald JZ, Pfenning LH, et al. The phoma-like dilemma. Stud Mycol. 2020;96:309–396. doi: 10.1016/j.simyco.2020.05.001.
- Database of plant diseases in Japan [Internet]. The Phytopathological Society of Japan; 2023 [cited 2023 Jul 25]. Available from: https://www.gene.affrc.go.jp/databases-micro_pl_diseases_en.php
- List of plant diseases in Korea [Internet]. Korean Society of Plant Pathology; 2023 [cited 2023 Jul 25]. Available from: http://genebank.rda.go.kr/kplant disease.do
- Park M-J, Lee H, Ryoo R, et al. A rapid and universal direct PCR method for macrofungi. Kor J Mycol. 2021;49:455–467.
- Rehner SA, Samuels GJ. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol Res. 1994;98(6):625–634. doi: 10.1016/S0953-7562(09)80409-7.
- Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172(8):4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990.
- De Hoog GS, Gerrits van den Ende AHG. Molecular diagnostics of clinical strains of filamentous Basidiomycetes. Mycoses. 1998;41(5–6):183–189. doi: 10.1111/j.1439-0507.1998.tb00321.x.
- White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, et al., editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. p. 315–322.
- Woudenberg JHC, Aveskamp MM, De Gruyter J, et al. Multiple Didymella teleomorphs are linked to the Phoma clematidina morphotype. Persoonia. 2009;22(1):56–62. doi: 10.3767/003158509X427808.
- Sung G-H, Sung J-M, Hywel-Jones NL, et al. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): identification of localized incongruence using a combinational bootstrap approach. Mol Phylogenet Evol. 2007;44(3):1204–1223. doi: 10.1016/j.ympev.2007.03.011.
- Liu YJ, Whelen S, Hall BD. Phylogenetic relationships among Ascomycetes: evidence from an RNA polymerase II subunit. Mol Phylogenet Evol. 1999;16(12):1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092.
- Chen Q, Bakhshi M, Balci Y, et al. Genera of phytopathogenic fungi: GOPHY 4. Stud Mycol. 2022;101(1):417–564. doi: 10.3114/sim.2022.101.06.
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340.
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054.
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (*and other methods) Version 4.0a. Sunderland: Sinauer Associates; 2003.
- Nylander JAA. MrModeltest version 2.4. Uppsala: Evolutionary Biology Centre; 2004.
- Ronquist F, Teslenko M, van der Mark P, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model selection across a large model space. Syst Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029.
- Rambaut A. FigTree version 1.4.4. Edinburgh: Institute of Evolutionary Biology; 2018.
- De Gruyter J, Noordeloos ME, Boerema GH. Contributions towards a monograph of Phoma (Coelomycetes) – I. 2. Section Phoma: additional taxa with very small conidia and taxa with conidia up to 7 μm long. Persoonia. 1993;15:369–400.
- Chen Q, Zhang K, Zhang G, et al. A polyphasic approach to characterise two novel species of Phoma (Didymellaceae) from China. Phytotaxa. 2015;197(4):267–281. doi: 10.11646/phytotaxa.197.4.4.
