 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Hydnum is a genus of ectomycorrhizal fungi belonging to the Hydnaceae family. It is widely distributed across different regions of the world, including North America, Europe, and Asia; however, some of them showed disjunct distributions. In recent years, with the integration of molecular techniques, the taxonomy and classification of Hydnum have undergone several revisions and advancements. However, these changes have not yet been applied in the Republic of Korea. In this study, we conducted an integrated analysis combining the morphological and molecular analyses of 30 specimens collected over a period of approximately 10 years in the Republic of Korea. For molecular analysis, the sequence data of the internal transcribed spacer (ITS) region, the large subunit of nuclear ribosomal RNA gene (nrLSU), and a portion of translation elongation factor 1-α (TEF1) were employed as molecular markers. Through this study, we identified eight species that had previously not been reported to occur in the Republic of Korea, including one new species, Hydnum paucispinum. A taxonomic key and detailed descriptions of the eight Hydnum species are provided in this study.
1. Introduction
The genus Hydnum L. belongs to the family Hydnaceae [Citation1,Citation2], and its type species is H. repandum [Citation3]. This genus is characterized by pileate to stipitate basidiomata; aculeate hymenophores; cylindrical to clavate basidia; and subglobose to ovoid, elliptical, colorless basidiospores. The presence of an aculeate hymenophore is the most distinctive characteristic of Hydnum [Citation4–6]. Hydnum plays an important role in forest ecology, as it forms ectomycorrhizal relationships with certain trees, such as Pinaceae and Fagaceae [Citation7–9].
In the early fungal taxonomic generation, terrestrial agaric mushrooms with aculeate hymenophores were assigned to the genus Hydnum [Citation10–12]. As a result, there are currently 978 records of Hydnum species registered in the Index Fungorum (http://www.indexfungorum.org/, accessed on May 26, 2023). Sequence-based classification (SBC) and identification (SBI) methods have been widely used to overcome the limitations of morphology-based identification of fungi [Citation13]. Molecular phylogenetic analysis has shown that the aculeate hymenophore has evolved independently several times in distantly related taxa [Citation14]. Consequently, many species formerly classified as Hydnum have been reclassified into other genera such as Hydnellum and Sarcodon [Citation15,Citation16]. The SBC of Hydnum has undergone continuous revisions [Citation2,Citation17,Citation18], leading to the reestablishment of the Hydnaceae family, including the genus Hydnum [Citation19]. Furthermore, the SBI has revealed numerous misidentifications of species within the genus Hydnum [Citation20–22].
Currently, only one Hydnum species (H. repandum) and its varieties (H. repandum var. album) have been described in the Republic of Korea [Citation23]. Previously, six Hydnum species and one variety were reported (H. aspratum, H. caput-medusae, H. crinaceus, H. imbricatum, H. repandum, H. repandum var. album, H. septentrionale). However, many of these species were subsequently reclassified into other genera. Only reports of H. repandum and its variety H. repandum var. album have persisted in the Republic of Korea for over 80 years [Citation24]. Recent studies based on the SBC and SBI have revealed that many Asian mycorrhizal species have been misidentified, often using the names of European and American species [Citation25–27]. Hydnum repandum and H. repandum var. album are ectomycorrhizal species named after European specimens; however, the applicability of these names has not been clarified through molecular analyses. Additionally, a previous analysis of global Hydnum diversity indicated clear differences between East Asian species and those from other continents [Citation17]. Therefore, taxonomic studies based on molecular phylogeny are necessary to elucidate mycorrhizal Hydnum species in the Republic of Korea.
To address taxonomic uncertainties and uncover the true diversity of Hydnum species in the Republic of Korea, we investigated hydnoid specimens preserved in three main Korean Fungariums. We employed three molecular markers: the sequences of internal transcribed spacer (ITS), nuclear ribosomal large subunit RNA (nrLSU), and the translation elongation factor 1-α (TEF1) gene. As a result, a total of eight Hydnum species were identified in the Republic of Korea. Among these, one species is potentially new to science, emphasizing the significance of this research in revealing the previously unrecognized Hydnum diversity in the Republic of Korea.
2. Materials and methods
2.1. Samples and morphological observations
Thirty Hydnum specimens, collected between 2009 and 2021, were examined (). The specimens were sourced from three institutions: the Seoul National University Fungus Collection (SFC), the Korea National Arboretum (KA), and the National Institute of Biological Resources of Korea (NIBR). Most specimens were collected within the Republic of Korea, while a few were obtained from Central Siberia, Russia and Lạc Dương District, Vietnam. The detailed collection information for each specimen is presented in . Most of the specimens were temporarily identified as H. repandum based on their morphology. Specimen information such as locality, habitat, and photographs was obtained from each fungarium.
Table 1. Summary of information information of the Hydnum specimens collected for the present study.
Macromorphological features of fresh basidiomata were described based on photographs of fresh specimens taken in the field. Color names and alphanumeric codes were rated using the Methuen Handbook of Colour [Citation28]. Micromorphological features, including basidiospores, basidia, and hyphae, were observed and measured in dried specimens using an Eclipse 80i compound light microscope (Nikon, Japan) and NIS ELEMENT BR software v3.2 (Nikon, Japan). Small pieces of spinose structures from each specimen were mounted in 5% KOH and stained with 1% Congo red. A minimum of 20 basidia, 30 basidiospores, and 20 other microstructures were measured for each specimen. The measurements were taken at 400× magnification using ImageJ software [Citation29]. The E value represents the length-to-width ratio of basidiospores, and the Q value indicates the average E value. The Q value was used to determine the shape of the spore morphology, and the range of E values and the photo taken during microscopic observation were used to confirm this using A Glossary of Mycology [Citation30]. Melzer’s reagent was used to assess the amyloid reaction [Citation31].
2.2. Molecular approaches: DNA extraction, PCR, sequencing, and phylogenetic analysis
The tissue samples were placed in CTAB buffer and homogenized using a bead ruptor elite homogenizer (OMNI International, Kennesaw, GA, USA). Genomic DNA was extracted using the AccuPrep Genomic DNA Extraction Kit (Bioneer Co., Daejeon, Korea) according to the manufacturer’s protocol. The ITS region was amplified using the primer set ITS1F/ITS4 [Citation32,Citation33] and the nrLSU region was amplified using three different primer sets (LR0R/LR3, LR5, and LR7) [Citation33]. For the TEF1 region, Hydnum-specific primers from previous studies were used (HEF1F/HEF1R and HydTEF1-F/HydTEF1-R) [Citation17,Citation34]. Polymerase chain reaction was performed using the AccuPower PCR premix (Bioneer Co., Daejeon, Korea) in a C1000 thermal cycler (Bio-Rad, Richmond, CA, USA), according to the PCR conditions described by [Citation26]. The PCR amplicons were verified by electrophoresis on a 1% agarose gel and purified using an Expin PCR Purification Kit (GeneAll Biotechnology, Seoul, Korea). The same primer pairs were used for sequencing, which was performed using an ABI Prism 3700 Genetic Analyzer (Life Technologies, Gaithersburg, MD, USA) by Macrogen (Seoul, Republic of Korea).
The generated sequences were manually proofread and edited using MEGA7 [Citation35] and deposited in GenBank. The aligned sequences were checked and edited manually using MEGA7 [Citation35]. For the phylogenetic analysis, reference sequences were retrieved from GenBank and UNITE and analyzed using our sequences (). Although the TEF1 region has recently been used in Hydnum phylogenetic studies [Citation17,Citation19], there are currently not enough TEF1 region sequences available for phylogenetic analysis. Therefore, this study provided supplemental TEF1 sequence information for Korean specimens to aid future research. All sequences of the ITS and nrLSU regions were aligned using MAFFT v7 [Citation36] and the alignments were manually edited using Geneious (https://www.geneious.com). For ITS phylogeny, three Sistotrema sequences were selected as the outgroup [Citation19]. For Maximum Likelihood (ML) analysis utilizing the combined ITS and nrLSU sequence datasets, only specimens that possessed both ITS and nrLSU sequences were included in the analysis. The sequences of two Sistotrema specimens possessing both ITS and nrLSU sequences were selected as outgroups. The final alignment was generated as a concatemer by manually merging the aligned sequences of two regions (ITS and nrLSU). Maximum Likelihood analysis was performed using RAxML–HPC v8 and a GTR + GAMMA model with 1,000 bootstrap [Citation37].
Table 2. Summary of information about the published Hydnum sequences used in the phylogenetic analysis.
3. Results
3.1. Phylogenetic analysis
We generated 76 sequences from 30 specimens, comprising 30 ITS, 28 nrLSU, and 18 TEF1 sequences. Since many reference sequences of Hydnum species were available only for the ITS region, we conducted ML analysis using only the sequence of the ITS region. In total, 144 ITS sequences, including three sequences of Sistotrema as an outgroup, were used. In the phylogenetic tree, Hydnum was divided into six subgenera, consistent with previous studies [Citation6,Citation19], and each subgenus was supported by a high bootstrap value, except for the subgenus Alba s. l.
Thirty Hydnum specimens were classified into eight taxa in the phylogenetic tree. They formed distinct clades with their corresponding reference sequences supported by high bootstrap values (except for the new species candidate) in four subgenera: Alba, Alba s. l., Pallida, and Rufescentia (). Seven of the taxa were identified as described species and one was identified as a new species candidate. In the subgenus Alba, two species were identified with four specimens: H. cremeoalbum (SFC20181012-04) and H. pinicola (KA13-1480, SFC20170811-18, and SFC20180928-18). In the subgenus Alba s. l., 11 specimens were assigned to two species, H. orientalbidum (SFC20150902-97, NIBRFG0000109936, NIBRFG0000113465, and NIBRFG0000122620) and H. minus (KA13-1475, KA16-1189, KA17-0894, KA20-0486, KA20-0682, KA20-0770, and KA20-0815), whereas one specimen (KA20-0732) was not identified at the species level. In subgenus Pallida, three specimens (KA16-0264, KA16-0511, and SFC20180705-81) were identified as H. pallidomarginatum and two specimens (KA16-0626 and KA16-1082) were identified as H. albopallidum. Within the subgenus Rufescentia, six specimens (KA16-0826, NIBRFG0000502952, NIBRFG0000508910, NIBRFG0000508951, NIBRFG0000509012, and SFC20210902-27) were identified as H. ventricosum. The sequences of three other specimens (NIBRFG0000503828, NIBRFG0000507153, and NIBRFG0000508573) did not match any of the described Hydnum species and formed a distinct monophyletic clade in the section Magnorufesentia in the subgenus Rufescentia, suggesting that they represent a new species candidate. To confirm the robust phylogenetic placement of these species, a phylogenetic analysis was conducted for species with both ITS and nrLSU sequences. All species identified in this study were supported by high bootstrap values. The morphological characteristics of eight Hydnum species are depicted in and , while their corresponding Korean names are suggested in Supporting Information Table S1.
Figure 1. Phylogenetic tree based on Maximum Likelihood (RAxML) analysis of Hydnum based on sequence data of ITS. Bootstrap values exceeding 70% are indicated at the branch nodes. Newly generated sequences from this study are represented in orange.
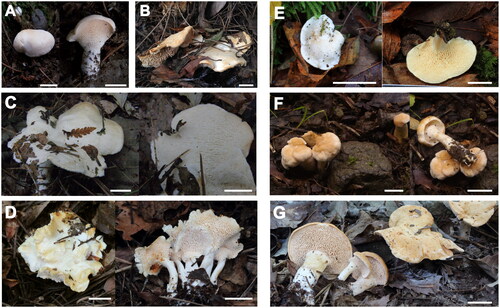
Figure 2. Pictures of basidiomata of seven Hydnum species from East Eurasia. (A) Hydnum cremeoalbum (SFC20181012-04); (B) H. pinicola (KA13-1480); (C) H. orientalbidum (NIBRFG0000113465); (D) H. minus (KA17-0894); (E) H. albopallidum (KA16-1082); (F) H. pallidomarginatum (SFC20180705-81); and (G) H. ventricosum (KA16-0826). Scale bars: 1 cm.
4. Taxonomy
4.1. Hydnum subgenus Alba
4.1.1. Hydnum cremeoalbum Liimat. & Niskanen, 2018 ()
Description. Pileus 20–30 mm in diameter, expanding to plane, often umbilicate, entire to irregular in outline; surface dull, dry, smooth, and glabrous, not zonate, whitish; margin incurved to straight, undulating, entire to obscurely lobed. Hymenophore slightly decurrent, 0.4–0.6 mm in length, 3–7 spines/mm2 crowded, concolorous with the pileus surface. Stipe cylindrical to subclavate, 16–18 × 7.5–9 mm, central or eccentric, solid; surface smooth, whitish, or concolorous with the pileus surface. Basidia narrowly clavate to narrowly urniform, 35–49 × 5–7 μm, with a basal clamp, producing (4–)5 sterigmata. Basidiospores subglobose to ellipsoid, 4.5–6.5 × 3.2–5.7 μm, E = 1.07–1.43, Q = 1.29, smooth, thin-walled, hyaline, sometimes with oil-drops, non-amyloid.
Habitat and distribution. Solitary, in a mixed forest of dominant with P. densiflora.
Specimens examined. SFC20181012-04, Hongdo-ri, Heuksan-myeon, Sinan-gun, Jeollanam-do, Republic of Korea, October 12, 2018, collected by Jae Young Park.
Remark. Hydnum cremeoalbum is characterized by the whitish pileus, cylindrical to subclavate spine, and 5–7 × 3.5–5.5 μm sized, ellipsoid basidiospores [Citation38]. The characteristics of the specimens used in this study mostly fell within the range of those described in the original description [Citation38]. The only difference observed was that the basidiomata size of the Korean Hydnum cremeoalbum was smaller than that in the original description (pileus, 30–70 mm; stipe, 25–35 × 15–30 mm) [Citation38].
4.1.2. Hydnum pinicola R. Sugaw. & N. Endo, 2022 ()
Description. Pileus 50–75 mm in diameter, plane, depressed; surface dull, dry, smooth, and not zonate, whitish to orange white (5A2); margin entire, incurved to straight. Hymenophore shortly decurrent, 2–4.5 mm in length, 4–6 spines/mm2 crowded, concolorous with the pileus surface. Stipe cylindrical to subclavate, 26–36 × 9.5–12.5 mm, central or eccentric, solid; surface smooth, concolorous with the pileus surface. Basidia (narrowly) utriform, 28–37 × 5.3–7.8 μm, with a basal clamp, producing 2–6 sterigmata. Basidiospores subglobose, 4.2–5.7 × 3.2–5 μm, E = 1.05–1.33, Q = 1.17, smooth, hyaline, sometimes with oil-drops, non-amyloid.
Habitat and distribution. Solitary or gregarious, in a mixed forest of dominant with P. densiflora.
Specimens examined. KA13-1480, Udu Mountain, Suwol-ri, Gajo-myeon, Geochang-gun, Gyeongsangnam-do Republic of Korea, July 17, 2013, collected by Sang Kuk Han; SFC20170811-18, Ordzhonikidzevskiy Rayon, Khakassia, Russia, August 11, 2017, collected by Young Woon Lim; SFC20180928-18, Deokgye-ri, Yeongdeok-gun, Gyeongsangbuk-do, Republic of Korea, collected by Young Woon Lim, Ki Hyeong Park, Abel.
Remark. Most features were consistent with the original description of Hydnum pinicola in Japan [Citation34]. Despite the geographical distance, the specimen (SFC20170811-18) collected in Central Siberia shared the same habitat and DNA sequence as those collected in East Asia.
4.2. Hydnum subgenus Alba s. l
4.2.1. Hydnum orientalbidum R. Sugaw. & N. Endo, 2022 ()
Description. Pileus 11–45 mm in diameter, expanding to pulvinate, often plane, entire to irregular in outline; surface dull, dry, smooth, not zonate, whitish; margin incurved to decurved, entire. Hymenophore shortly decurrent, 1–4 mm in length, 4–6 spines/mm2 crowded, concolorous with the pileus surface. Stipe subclavate, 15–30 × 5.5–13 mm, central or eccentric, solid; surface smooth, concolorous with the pileus surface. Basidia urniform to utriform, 28–38.2 × 5.2–8 μm, with a basal clamp, producing 3–6 sterigmata. Basidiospores subglobose, 4.5–5.6 × 4.1–5.3 μm, E = 1.00–1.36, Q = 1.15, smooth, hyaline, sometimes with oil-drops, non-amyloid.
Habitat and distribution. Solitary to gregarious, in a mixed forest.
Specimens examined. NIBRFG0000109936, Hakgok-ri, Socho-myeon, Wonju-si, Gangwon-do, Republic of Korea, 15 August 2009, collected by Young Woon Lim; NIBRFG0000113465, Unak Mountain, Hwahyeon-myeon, Pocheon-si, Gyeonggi-do, Republic of Korea, collected by Jae-Jin Kim, Yongil Kim, Yeongseon Jang; SFC20150902-97, Jajangnamusup-gil 760, Inje-eup, Inje-gun, Gangwon-do, Republic of Korea, September 2, 2015, collected by Young Woon Lim.
Remark. Hydnum orientalbidum has white basidiomata that resemble those of H. albidum. While the latter species has been reported in North America, the former has been confirmed to occur only in Asia and exhibits distinct differences in terms of its DNA sequence, as mentioned in previous study [Citation34]. The Korean H. orientalbidum specimen matched the original morphological, geographical, and ecological characteristics with the original description. However, the basidiomata of the studied specimens are generally small, particularly in the pileus and stipes [Citation34].
4.2.2. Hydnum minus Yanaga & N. Maek., [as “minum”], 2015 ()
Description. Pileus 16–38 mm in diameter, expanding to plane or sometimes concave, depressed to umbilicate, irregular in outline; surface dull, glabrous, slightly zonate in center, whitish to yellowish white (4A2) to pale orange (5A3); margin curved, becoming undulate in age, entire to obscurely lobed. Hymenophore spinose, decurrent; spines conical to aciculate, 0.78–1.7 mm in length, 6–11 spines/mm2 crowded, more buff (6A2 to 7A3) than the pileus surface. Stipe cylindrical to obclavate, 15–20 2–4 mm, central or eccentric; surface smooth, concolorous with the pileus surface. Basidia subclavate to suburniform, constricted, 25.5–32.46 (35)
4.8–6.7
with a basal clamp, producing 4–6 sterigmata. Basidiospores subglobose to ellipsoid, 4–5.3 × 2.6–3.5
E = 1.25–1.6, Q = 1.47, smooth, thin-walled, hyaline, with oil-drops, non-amyloid.
Habitat and distribution. Solitary to gregarious, in a mixed forest.
Specimens examined. KA13-1475, Udu Mountain, Suwol-ri, Gajo-myeon, Geochang-gun, Gyeongsangnam-do, Republic of Korea, October 17, 2013, collected by Sang Kuk Han; KA16-1189, Sogwang-ri, Seo-myeon, Ulleung-gun, Gyeongsangbuk-do, Republic of Korea, October 5, 2016, collected by Sang Kuk Han; KA17-0894, Sogwang-ri, Seo-myeon, Ulleung-gun, Gyeongsangbuk-do, September 14, 2017, collected by Jong Won Jo, Yeongnam Gwak, Juyeong Park, Huisu Jeong; KA20-0486, Dongmyeon-ri, Jaesan-myeon, Bonghwa-gun, Gyeongsangbuk-do, Republic of Korea, August 13, 2020, collected by Jong Won Jo, Hyun Lee, Hyeongso Kim, Sangyeong Park; KA20-0682, Dongmyeon-ri, Jaesan-myeon, Bonghwa-gun, Gyeongsangbuk-do, Republic of Korea, September 14, 2020, collected by Jong Won Jo, Hyeongso Kim, Sangyeong Park; KA20-0770, Doota Mountain, Miro-myeon, Samcheok-si, Gangwon-do, Republic of Korea, September 16, 2020, collected by Jong Won Jo, Hyeongso Kim, Sangyeong Park; KA20-0815, 1610, Guryongnyeong-ro, Seo-myeon, Yangyang-gun, Gangwon-do, Republic of Korea, September 17, 2020, collected by Jong Won Jo, Hyeongso Kim, Sangyeong Park.
Remark. All specimens were collected in the Republic of Korea, and the Korean Hydnum minus has a wider pileus diameter range, wider and longer stipe, and slightly smaller basidiospores when compared with the reference (pileus 10–25 mm in diameter; stipe 8–15 × 2–5 mm in width; basidiospores 4.5–5.5 × 3–4.5 μm) [Citation38]. The epithet “minus (minum)” was originated from the small basidiomata of the holotype, but the Korean Hydnum minus has a greater pileus width range compared to that of the original description. Furthermore, the phylogenetic tree revealed intraspecific sequence variation in Hydnum minus (), which was within the acceptable range for this species (less than 0.05%).
4.3. Hydnum subgenus Pallida
4.3.1. Hydnum albopallidum R. Sugaw. & N. Endo, 2022 ()
Description. Pileus 8–30 mm in diameter, expanding to plane, pulvinate; surface dull, glabrous, slightly zonate in center, whitish to orange white (5A2); margin curved, entire. Hymenophore spinose, decurrent; spines 0.5–0.8 mm in length, 6–8 spines/mm2 crowded, concolorous with the pileus surface. Stipe cylindrical, 8–153–7.3 mm, central or slightly eccentric; surface smooth, concolorous with the pileus surface. Basidia urniform to (narrowly) utriform, sometimes clavate, 37.2–53.8 (56)
8.5–11.4
with a basal clamp, producing 3–4 sterigmata. Basidiospores subglobose, 7.1–8.7 × 6.2–8
E = 1.03–1.29, Q = 1.11, smooth, thin-walled, hyaline, with oil-drops, non-amyloid.
Habitat and distribution. Solitary, in a mixed forest, nearby the bryophyte.
Specimens examined. KA16-0626, Eorimok Valley, Haean-dong, Jeju-si, Jeju Island, Republic of Korea, July 18, 2016, collected by Sang Kuk Han; KA16-1082, Eorimok Valley, Haean-dong, Jeju-si, Jeju Island, Republic of Korea, September 26, 2016, collected by Sang Kuk Han.
Remark. Overall, the characteristics of these specimens matched the original description; however, their basidiospores were slightly smaller than those of the original description (8–9.5 × 7–8 ) [Citation34].
4.3.2. Hydnum pallidomarginatum T. Cao & H. S. Yuan., 2021 ()
Description. Pileus 10–42.9 mm in diameter, plane, depressed in center; surface dull, dry, smooth, and not zonate, orange white (5A2) to light orange (5A4); margin entire, incurved to straight, whitish. Hymenophore subdecurrent, 2–5 mm in length, 4–7 spines/mm2 crowded, white to orange white (5A2). Stipe cylindrical, 26–36 × 8.5–10 mm, central, solid; surface smooth, concolorous with the hymenophore. Basidia urniform to utriform, 32–55 × 5–12 μm, with a basal clamp, producing 2–4 sterigmata. Basidiospores ellipsoid, 7.4–10.1 × 6.3–7.9 μm, E = 1.27–1.38, Q = 1.31, smooth, hyaline, sometimes with oil-drops, non-amyloid.
Habitat and distribution. Solitary to gregarious, in a mixed forest.
Specimens examined. KA16-0264, Bakji island, 209, Bakjido-gil, Anjwa-myeon, Sinan-gun, Jeollanam-do, Republic of Korea, June 28, 2016, collected by Sang Kuk Han; KA16-0511, Bakji island, 209, Bakjido-gil, Anjwa-myeon, Sinan-gun, Jeollanam-do, Republic of Korea, July 12, 2016, collected by Sang Kuk Han; SFC20180928-18, Duryunsan, Samsan-myeon, Haenam-gun, Jeollanam-do, Republic of Korea, July 05, 2018, collected by Hae Jin Cho, Ki Hyeong Park, Dohye Kim.
Remark. Korean basidiomata have prominent features present in the original description, including a brighter pileus margin, an orange whitish to light orange pileus, and broadly ellipsoid basidiospores [Citation19]. However, the Korean H. pallidomarginatum has a larger pileus when comparing to the original description (pileus 20–35 mm in diameter) [Citation19]. In the phylogenetic tree, H. pallidomarginatum exists in the same clade as H. flabellatum, which can be distinguished based on the scabrous texture of the pileus and longer stipe [Citation19]. The morphological differences were subtle and did not show significant distinctions, suggesting that further research is needed to establish the criteria for the delimitation between these two species.
4.3.3. Hydnum ventricosum Cao & H.S. Yuan, 2021 ()
Description. Pileus 20–55 mm in diameter, convex to plane, occasionally depressed in the center; surface dull, dry, smooth, and glabrous, not zonate, pale ocher, occasionally orangish ocher with whitish at the margin; margin entire. Hymenophore spinose, adnate, decurrent; spines conical to aciculate, 1.2–5.4 mm in length, 2–7 spines/mm2 crowded, concolorous with the stipe surface. Stipe cylindrical to subclavate, 29–69 7–19 mm, central or eccentric, solid; surface smooth, concolorous with the pileus surface. Basidia clavate to narrowly urniform, 33–51
8.3–12 μm, with a basal clamp, producing 3–4 sterigmata. Basidiospores globose, 6.8–8.6
6.5–8.4 μm, E = 1.01–1.05, Q = 1.03, smooth, thin-walled, hyaline, sometimes with oil-drops, non-amyloid.
Habitat and distribution. Solitary or in groups, occasionally gregarious, in a pile of leaves of mixed deciduous forest.
Specimens examined. KA16-0826, 415, Gwangneungsumogwon-ro, Soheul-eup, Pocheon-si, Gyeonggi-do, Republic of Korea, July 28, 2016, collected by Sang Kuk Han; NIBRFG0000509012, Taebaek Mountain, Gohan-eup, Jeongseon-gun, Gangwon-do, Republic of Korea, September 10, 2020, collected by Changmu Kim, Minkyeong Kim, Chorong Ahn; SFC20210902-27, Taebaek Mountain, Gohan-eup, Jeongseon-gun, Gangwon-do, Republic of Korea, September 2, 2021, collected by Young Woon Lim, Ki Hyeong Park, Shinnam Yoo, Yoonhee Cho.
Remark. Hydnum ventricosum is characterized by an orangish pileus, fusiform to subcylindrical basidia, and subglobose basidiospores [Citation19]. Korean H. ventricosum has a relatively larger pileus than original description (up to 35 mm) [Citation19].
4.3.4. Hydnum paucispinum JS Kim, C Kim, and YW Lim, sp. nov. () MycoBank no.: MB#849344
Holotype. NIBRFG0000503828, Đạ Chais, Lạc Dương, Lâm Đồng, Vietnam, June 16, 2018, collected by Changmu Kim, Jae Young Park, Jin Sung Lee, in mixed forest dominant with P. densiflora. GenBank: ITS = OR211379; nrLSU = OR211395; TEF1 = OR220062.
Etymology. Paucispinum, refers to the relatively less crowded aculeate hymenophore (paucus means fewer, and spinum refers from aculeate hymenophore)
Diagnosis. Hydnum paucispinum is characterized by whitish to light orange pileus, a non-decurrent hymenophore, cylindrical stipe, and 7.1–9.2 6.4–8.6 μm sized, subglobose basidiospores. Ecologically, this species occurs in solitary or gregarious forms in Pinaceae forests.
Pileus 15–35 mm in diameter, convex to plane, irregularly round, depressed in the center; surface dry, smooth, glabrous, zonate; pale orange (5A4) when dry, light orange (5A5 to 6A5) when wet; margin entire, arched when young. Hymenophore spinose, non-decurrent; spines conical, aculeate, 1.1–3 mm in length, 3–5 spines/mm2 crowded, concolorous with the pileus surface. Stipe cylindrical, 15–32 2.3–7 mm, central or slightly eccentric, solid; surface smooth, whitish.
Tramal hyphae of the spines 3–5.5 μm in diameter, smooth, thin-walled. Pileipellis an indefinite trichodermium, composed of hyphae similar to those of the tramal hyphae of the spine, 4–7.5 μm in diameter. Basidia narrowly utriform, 37–47.5 8.1–11.4 μm, with a basal clamp, producing 2–4 sterigmata. Basidiospores subglobose, 7.1–9.2
6.4–8.6 μm, E = 1.00–1.16, Q = 1.06, smooth, thin-walled, hyaline, sometimes with oil-drops, non-amyloid.
Habitat and distribution. Solitary or gregarious, in a mixed forest dominant with P. densiflora.
Additional specimens examined. NIBRFG0000507153, Xã Đạ Nhim, Lạc Dương, Lâm Đồng, Vietnam, July 1, 2019, collected by Changmu Kim, Jin Sung Lee, Jae Young Park, Nam Kyu Kim; NIBRFG0000508573, Gaya Mountain, Gaya-myeon, Hapcheon-gun, Gyeongsangnam-do, Republic of Korea, July 1, 2019, collected by Changmu Kim.
Remark. The morphological features of Hydnum paucispinum match the representative features of the subgenus Rufescentia [Citation6]. This species belongs to the Magnorufescentia section within this subgenus based on the Q value and morphology of basidiospores, which are characteristics that differentiate the two sections in this subgenus [Citation6]. Hydnum paucispinum differs from H. ventricosum in several aspects, including having shorter spines (1.1–3 mm vs. 1–5 mm long), a relatively sparse density of hymenophore (1–2 spines per mm vs. 2–4 spines per mm), a shorter and slender stipe (15–32 2.3–7 mm vs. 30–35
10–15 mm), narrowly utriform basidia, and a slightly thinner pileipellis (4–7.5 μm vs. 5–10 μm) [Citation19].
5. Taxonomic key of Korean Hydnum species
1 Pileus white to pale yellow 2
1 Pileus pale orange to reddish orange (sometimes whitish) 5
2 Spines less crowded (≤6 spines/mm2) 3
2 Spines crowded (≥6 spines/mm2) Hydnum minus
3 The length of hymenophore > 1 mm 4
3 The length of hymenophore < 1 mm H. cremeoalbum
4 Brownish discoloration of the stipe* H. orientialbidum
4 Lack of discoloration of the stipe* H. pinicola
5 Spines crowded (≥5 spines/mm2 on average) 6
5 Spines less crowded (≤5 spines/mm2) H. paucispinum
6 Q value of basidiospores < 1.2 7
6 Q value of basidiospores > 1.2 H. pallidomarginatum
7 The length of hymenophore> 1 H. ventricosum
7 The length of hymenophore < 1 H. albopallidum
*This characteristic was not described in this study but was derived from the reference paper [Citation9].
5. Discussion
An integrative study combining morphological and molecular analyses confirmed the presence of eight Hydnum species in the Republic of Korea. Previously, all the specimens deposited in the three Korean fungaria were identified as Hydnum repandum. However, Hydnum repandum, the type species of Hydnum reported in Europe [Citation6,Citation39], seems absent from the Republic of Korea. The eight Hydnum species identified have aculeate hymenophores, which may lead to confusion with H. repandum. Nevertheless, as demonstrated in the taxonomic key of this study, closer examination revealed distinct morphological differences. Additionally, the sequences of these eight species were distinct from H. repandum.
Based on the results of phylogenetic and morphological analyses, seven of the eight Hydnum species were identified as described species. In the phylogenetic analysis, all species formed well-resolved clusters with the reference sequences of each species, supported by high bootstrap values, and no significant differences were observed when compared to the original descriptions. However, certain species exhibited variations in the size of their basidiomata compared with the original descriptions. These differences may be attributed to variations in the growth of basidiomata in different climatic zones, or to the limited number of specimens. Therefore, to generalize the characteristics of the Korean Hydnum, further examination using a larger number of specimens is necessary. In addition, further research on interspecies delimitation is required for some species. For example, H. ventricosum formed a monophyletic clade with H. subberkelyanum, showing no genetic variation in the ITS region between them. Their morphological characteristics were almost identical except for the width of the stipipellis [Citation19,Citation34]. Therefore, H. subberkeleyanum should be synonymized with H. ventricosum, as the latter species was proposed earlier.
Consistent with previous studies, taxonomic investigations of fungal specimens collected in Asia differed considerably from those conducted in Western regions. In the case of Hydnum, an ectomycorrhizal fungus, the species distribution may vary depending on the distribution of the host plants [Citation40,Citation41]. The disjunct distributions of certain endemic species have been reported in Hydnum [Citation17]. Due to this reason, numerous new species have been proposed in recent taxonomic studies in Asia. The seven described species identified in the Republic of Korea have been recorded exclusively in Asia and not on any other continents [Citation6,Citation17,Citation19,Citation38]. Additionally, the new species proposed in this study, Hydnum paucispinum, was found in the Republic of Korea and the high mountains of Vietnam, and its molecular distinctiveness from other described species was clearly shown. Our results indicate that despite the distance between East Asia, the mountainous areas of Vietnam, and central Siberia, their Hydnum species share similar ecological traits. These findings support those of previous studies highlighting the differences between Asian ectomycorrhizal fungi and those in western regions, which are attributed to variations in continents and ecological environments [Citation42–44]. Therefore, continued research on the fungal taxonomy in the eastern region provides an opportunity to further explore the diversity of fungi, which has only been partially revealed thus far [Citation45].
In conclusion, the present study confirmed the presence of eight Hydnum species in the Republic of Korea using SBI and morphological analyses. None of these species correspond to Hydnum repandum. Given the similarity in morphology among certain Hydnum species, careful observation of microscopic features and SBI processes are essential to prevent misidentification. The findings of this study will enhance accurate identification of Hydnum species in the Republic of Korea by providing validated sequence data and comprehensive morphological characteristics.
Supplemental Material
Download MS Word (12 KB)Supplemental Material
Download MS Excel (11.1 KB)Supplemental Material
Download JPEG Image (1.1 MB)Acknowledgment
We would like to thank Editage (www.editage.co.kr) for English language editing.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data generated in this study are all included in the article, and the sequence data have been deposited in GenBank.
Additional information
Funding
References
- Miller LW. The genera of Hydnaceae. Mycologia. 1933;25(4):286–302. doi: 10.1080/00275514.1933.12020669.
- Moncalvo JM, Nilsson RH, Koster B, et al. The cantharelloid clade: dealing with incongruent gene trees and phylogenetic reconstruction methods. Mycologia. 2006;98(6):937–948. doi: 10.3852/mycologia.98.6.937.
- Petersen RH. The typification of Hydnum L. ex Fries. Taxon. 1973;22(1):99–104. doi: 10.2307/1218039.
- Donk M. The generic names proposed for Hymenomycetes. V, "Hydnaceae" (continuation). Taxon. 1956;5(5):95–115. doi: 10.2307/1216240.
- Maas Geesteranus R. Hydnaceous fungi of the Eastern old world. Vehr K Ned Akad Wet. 1971;II.60:1–176.
- Niskanen T, Liimatainen K, Nuytinck J, et al. Identifying and naming the currently known diversity of the genus Hydnum, with an emphasis on European and North American taxa. Mycologia. 2018;110(5):890–918. doi: 10.1080/00275514.2018.1477004.
- Diamandis S, Perlerou C. The mycoflora of the chestnut ecosystems in Greece. For Snow and Landscape Res. 2001;76(3):499–504.
- Raidl S, Agerer R. Studien an ektomykorrhizen. XLII: ontogenie der rhizomorphen von Laccaria amethystina, Hydnum rufescens und Sarcodon imbricatus. Nova Hedwigia. 1992;55(3–4):279–307.
- Sugawara R, Sotome K, Maekawa N, et al. Mycorrhizal synthesis, morpho-anatomical characterization of mycorrhizae, and evaluation of mycorrhiza-forming ability of Hydnum albidum–like species using monokaryotic and dikaryotic cultures. Mycorrhiza. 2021;31(3):349–359. doi: 10.1007/s00572-021-01024-7.
- Harrison KA. New or little known North American stipitate Hydnums. Can J Bot. 1964;42(9):1205–1233. doi: 10.1139/b64-116.
- Maas Geesteranus RA. Notes on hydnums. Persoonia-Molecular Phylogeny and Evolution of Fungi. 1960;1(3):341–384.
- Murrill WA. Additions to Florida fungi-V. Vol. 67(4), In: Bulletin of the Torrey Botanical Club; New York (NY): Torrey Botanical Society; 1940. p. 275–281. doi: 10.2307/2481174.
- Grigoriev IV, Cullen D, Goodwin SB, et al. Fueling the future with fungal genomics. Mycology. 2011;2(3):192–209.
- Hibbett DS, Pine EM, Langer E, et al. Evolution of gilled mushrooms and puffballs inferred from ribosomal DNA sequences. Proc Natl Acad Sci USA. 1997;94(22):12002–12006. doi: 10.1073/pnas.94.22.12002.
- Baird RE. Type studies of North American and other related taxa-of stipitate Hydnums. In: Bibliotheca Mycologica; 1986. ISBN 978-3-443-59004-8. [AQ]
- Larsson KH, Svantesson S, Miscevic D, et al. Reassessment of the generic limits for Hydnellum and Sarcodon (Thelephorales, Basidiomycota). MycoKeys. 2019;54:31–47. doi: 10.3897/mycokeys.54.35386.
- Feng B, Wang X-H, Ratkowsky D, et al. Multilocus phylogenetic analyses reveal unexpected abundant diversity and significant disjunct distribution pattern of the hedgehog mushrooms (Hydnum L.). Sci Rep. 2016;6(1):25586. doi: 10.1038/srep25586.
- Swenie RA, Baroni TJ, Matheny PB. Six new species and reports of Hydnum (Cantharellales) from Eastern North America. MycoKeys. 2018;42(42):35–72. doi: 10.3897/mycokeys.42.27369.
- Cao T, Hu Y-P, Yu J-R, et al. A phylogenetic overview of the Hydnaceae (Cantharellales, Basidiomycota) with new taxa from China. Stud Mycol. 2021;99(1):100121–100121.
- Huhtinen S, Ruotsalainen J. Variability of Hydnum rufescens in Finland: three taxa hidden under one name–and appearance? Karstenia. 2006;46(1):17–24. doi: 10.29203/ka.2006.412.
- Ostrow H, Beenken L. Hydnum ellipsosporum spec. nov. (Basidiomycetes, Cantharellales)–ein doppelgänger von Hydnum rufescens Fr. Zeitschrift Fuer Mykologie. 2004;70(2):137–156.
- Vizzini A, Picillo B, Ercole E, et al. Detecting the variability of Hydnum ovoideisporum (Agaricomycetes, Cantharellales) on the basis of Italian collections, and H. magnorufescens sp. nov. Mycosphere. 2012;4(1):32–44. doi: 10.5943/mycosphere/4/1/2.
- NIBR. National Institute of Biological Resources, 2022; [cited 2022 Mar 23]. Available from: https://kbr.go.kr/
- Kaburagi Y. Korea forest experiment station. Korean and Manchurian practical manual of forest. Tokyo: Yokendo; 1940. p. 339–367.
- Cho HJ, Park MS, Lee H, et al. A systematic revision of the ectomycorrhizal genus Laccaria from Korea. Mycologia. 2018;110(5):948–961.
- Lee H, Wissitrassameewong K, Park MS, et al. Taxonomic revision of the genus Lactarius (Russulales, Basidiomycota) in Korea. Fungal Divers. 2019;95(1):275–335. doi: 10.1007/s13225-019-00425-6.
- Wisitrassameewong K, Looney BP, Le HT, et al. Lactarius subgenus Russularia (Basidiomycota, Russulales): novel Asian species, worldwide phylogeny and evolutionary relationships. Fungal Biol. 2016;120(12):1554–1581. doi: 10.1016/j.funbio.2016.08.004.
- Kornerup A, Wanscher J. Methuen handbook of colour. London: Eyre Methuen Ltd; 1978. ISBN 978-0413334008.
- Collins TJ. ImageJ for microscopy. Biotechniques. 2007;43(1 Suppl):25–30. doi: 10.2144/000112517.
- Snell WH, Dick EA. A glossary of mycology. Cambridge (MA): Harvard University Press; 1971. ISBN 0674-35451-6.
- Largent DL, Stuntz DE. How to identify mushrooms to genus. 1973. ISBN: 978-0916422004.
- Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes‐application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2(2):113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x.
- White TJ, Bruns TD, Lee SB, et al. Analysis of phylogenetic relationships by amplification and direct sequencing of ribosomal RNA genes. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York: Academic Press; 1990. p. 315–322.
- Sugawara R, Maekawa N, Sotome K, et al. Systematic revision of Hydnum species in Japan. Mycologia. 2022;114(2):413–452. doi: 10.1080/00275514.2021.2024407.
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054.
- Katoh K, Misawa K, Kuma K, et al. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436.
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446.
- Yanaga K, Sotome K, Ushijima S, et al. Hydnum species producing whitish basidiomata in Japan. Mycoscience. 2015;56(4):434–442. doi: 10.1016/j.myc.2015.01.001.
- Linnaeus C. Species plantarum, exhibentes plantas rite cognitas ad genera relatas, cum differentiis specificis, nominibus trivialibus, synonymis selectis, locis natalibus, secundum systema sexuale digestas, Impensis GC Nauk; 1797. doi: 10.5962/bhl.title.37656.
- van der Linde S, Suz LM, Orme CDL, et al. Environment and host as large-scale controls of ectomycorrhizal fungi. Nature. 2018;558(7709):243–248. doi: 10.1038/s41586-018-0189-9.
- Yang H, Zang Y, Yuan Y, et al. Selectivity by host plants affects the distribution of arbuscular mycorrhizal fungi: evidence from ITS rDNA sequence metadata. BMC Evol Biol. 2012;12(1):50. doi: 10.1186/1471-2148-12-50.
- Cho HJ, Lee H, Park MS, et al. Two new species of Laccaria (Agaricales, Basidiomycota) from Korea. Mycobiology. 2020;48(4):288–295.
- Wilson AW, May TW, Mueller GM. Biogeography of the ectomycorrhizal mushroom genus Laccaria. In: Tedersoo L, editor. Biogeography of mycorrhizal symbiosis. ecological studies, Vol. 230. Cham: Springer; 2017. doi: 10.1007/978-3-319-56363-3_13.
- Wu BW, Gao C, Chen L, et al. Host phylogeny is a major determinant of Fagaceae-associated ectomycorrhizal fungal community assembly at a regional scale. Front Microbiol. 2018;9:2409. doi: 10.3389/fmicb.2018.02409.
- Phukhamsakda C, Nilsson RH, Bhunjun CS, et al. The numbers of fungi: contributions from traditional taxonomic studies and challenges of metabarcoding. Fungal Diver. 2022;114(1):327–386. doi: 10.1007/s13225-022-00502-3.

