Abstract
Phytophthora species, classified under Oomycota, cause significant damage to various crops and trees. The present study introduced Phytophthora species, P. nagaii and P. tentaculata, new to Korea, which pose notable risks to their respective host plants. Our research provided a comprehensive description of these species taking into account their cultural features, morphological characteristics, and molecular phylogenetic analysis using the internal transcribed spacer rDNA region and cytochrome c oxidase subunit mtDNA genes (cox1 and cox2) sequences. In addition, this study first evaluated the sensitivity of P. nagaii and P. tentaculata to five anti-oomycete fungicides, finding both species most responsive to picarbutrazox and P. tentaculata resistant to fluazinam. The data can guide targeted treatment strategies and offer insights into effective control methods. The findings expand our understanding of the diversity, distribution, and management of Phytophthora species in Korea.
1. Introduction
The genus Phytophthora, infamously called the “plant destroyer,” is a significant group of the phylum Oomycota. It was previously classified under the kingdom Fungi but is now recognized as a fungal-like member of the Kingdom Chromista. This genus is remarkably diverse, with over 200 known species [Citation1,Citation2]. This significant increase from an earlier estimate of around 120 species [Citation3] is attributed to the advent of molecular phylogenetic analysis.
Phytophthora species are renowned for causing critical diseases in a broad spectrum of agriculturally and ornamentally valuable crops and forest trees, with frequent reports from nurseries [Citation4,Citation5]. In addition, numerous species are featuring prominently in lists of global emergent threats to natural ecosystems [Citation6–11].
In Korea, considerable research efforts have been invested in Phytophthora species due to their profound impact on agricultural and ecological settings. As it stands, 22 Phytophthora species, including P. capsici, P. infestans, P. nicotianae, P. palmivora, and P. sojae, have been recognized [Citation12,Citation13], most species of which have been reported as disease-causing agents in diverse plants [Citation14–23]. The pioneering work in Phytophthora species identification in Korea began with the cataloguing and morphological examination of various species [Citation24], providing an in-depth account of disease etiology, epidemiology, and management, but also greatly enhancing our understanding of Phytophthora diseases in Korean agriculture. In recent years, the scope of studies has broadened to encompass molecular identification and phylogenetic analysis. Seo et al. [Citation25] conducted a molecular phylogenetic analysis for Phytophthora species in Korea, enriching our understanding of their genetic diversity and relationships. Moreover, a next-generation sequencing (NGS) investigation demonstrated that conventional farms employing chemical fertilizers and pesticides displayed a significantly higher abundance of plant pathogens, such as Phytophthora species, compared to organic farms [Citation26]. In addition, Phytophthora species were frequently found in freshwater environments, displaying a distinct preference for plant debris [Citation27].
In 2019, the Korean government implemented the Positive List System (PLS) to regulate the improper use of pesticides. This system specifies which pesticides suit particular plants and their corresponding diseases. As a result, this requires reevaluating how plant pathogen strains react to the range of fungicides used within the nation, including tests to gauge their resistance or sensitivity. Given this context, it is essential to examine how oomycete plant pathogens, such as Phytophthora species, react to commonly employed anti-oomycete fungicides. Currently, five fungicides, including metalaxyl, ethaboxam, dimethomorph, fluazinam, and picarbutrazox, are extensively applied in both national and international agricultural systems to control oomycete-related diseases [Citation28–36].
Metalaxyl suppresses the activity of ribosomal RNA synthetase in oomycete pathogens [Citation37,Citation38]. Ethaboxam targets foliar diseases caused by Phytophthora infestans on pepper and potato and works by disrupting the assembly of microtubulin [Citation31,Citation32,Citation39]. Dimethomorph hinders the enzyme responsible for cellulose synthesis, whereas Fluazinam, a member of the 2,6-dinitroanilines group, is broadly effective against various pathogens, including P. infestans [Citation40–42]. Picarbutrazox specializes in oomycetes but has an as-yet unidentified mechanism of action. Despite their effectiveness, these compounds can lead to the development of resistance in some cases. For instance, resistance risks have been documented with metalaxyl [Citation43,Citation44] and dimethomorph [Citation45]. As a result, ongoing monitoring and research are essential to ensure that these treatments remain effective in managing diseases caused by the target pathogens.
In the current research, Phytophthora isolates were identified by morphological and molecular phylogenetic methods and assessed their activities against five anti-oomycete fungicides. We also discussed their potential risks in domestic agriculture.
2. Materials and methods
2.1. Oomycete isolates
Four Phytophthora isolates used in this study were obtained from the Korean agricultural culture collection (KACC; Jeonju, Korea) (). They were cultured on V8 agar (V8A), composed of 200 mL clarified V8 juice, 10 g CaCO3, 15 g agar, 800 mL deionized water, and incubated at 25 °C for a week.
Table 1. Information of Phytophthora isolates used in this study.
2.2. Cultural and morphological analysis
To examine the colony growth patterns on three different media, a 4-mm agar plug was placed on plates of potato dextrose agar (PDA; Difco, Detroit, MI, USA), 20% V8A, and corn meal agar (CMA; Difco, Detroit, MI, USA). For observing the sexual organs of Phytophthora isolates (KACC 45737, 40909, 40912, and 40913), soil extract was utilized. The soil was moistened, set aside overnight, and subsequently filtered through Whatman No. 1 filter paper. The filtered soil extracts were then equally divided, and one portion passed through a 0.22 µm Millipore membrane. One-day-old mycelial plugs of Phytophthora isolates, grown in V8 liquid media, were rinsed with sterile deionized water and placed in a petri dish containing 10 mL of the soil extract. These plates were kept at room temperature (25 °C) and inspected at 2, 5, and 7 days.
The morphological traits of sporangia of KACC 40909, 40912, and 40913 were observed in the petri dish containing soil extract, as previously outlined. To stimulate the formation of sporangia in KACC 45737, an autoclaved leaf blade of Zoysia japonica was placed on a V8A medium, pre-inoculated with the KACC 45737 three days beforehand [Citation46]. After three days, the colonized leaf was moved to a new Petri dish filled with distilled water [Citation47]. All microscopic structures were examined using a Zeiss Axio Imager A2 microscope (Carl Zeiss, Oberkochen, Germany) and captured using a Dhyana 400DC camera (Tucsen, Fuzhou, China) attached to the microscope.
2.3. Molecular phylogenetic analysis
Genomic DNA was extracted with the MagListo 5 M Plant Genomic DNA Extraction Kit (Bioneer, Daejeon, Korea). The internal transcribed spacer (ITS) rDNA regions were amplified using the primer pairs ITS1/ITS4 [Citation48]. In addition, the cytochrome c oxidase subunit I (cox1) and cytochrome c oxidase subunit II (cox2) mtDNA of oomycete strains were amplified using OomCox1-levup/OomCox1-levlo [Citation49] and cox2-F [Citation50]/cox2-RC4 [Citation51], respectively. The PCR amplicons were purified using an AccuPrep PCR Purification Kit (Bioneer, Daejeon, Korea) and then sequenced by Macrogen Inc. (Seoul, Korea). Editing was carried out with DNAStar software package 5.05 (DNAStar, Inc., Madison, WI), followed by BLASTn search against the NCBI GenBank database.
Phylogenetic analysis was undertaken based on a concatenated dataset of ITS, cox1, and cox2 sequences, including reference sequences retrieved from NCBI GenBank, and aligned by the G-INS-i algorithm of MAFFT 7 [Citation52]. Phylogenetic trees were constructed using MEGA X with maximum likelihood (ML) and minimum evolution (ME) inferences, applying the Tamura-Nei model and bootstrapped with 1000 replicates.
2.4. Fungicide sensitivities
Anti-oomycete fungicides used in this study include metalaxyl (ai 25% WP), ethaboxam (ai 15% WP), dimethomorph (ai 50% WP), fluazinam (ai 25% WP), and picarbutrazox (ai 10% WP) (). The sensitivity of mycelial growth of Phytophthora isolates to the fungicides was tested using an agar dilution method. Each fungicide was dissolved in sterilized distilled water and then to amended to 20% V8A to obtain final concentrations of 0 (control), 0.01, 0.1, 1, 10, 100, and 1000 μg/mL. Rifampicin was added at a concentration of 15 μg/mL to avoid medium contamination. After culturing the pathogen on V8A for one week at 25 °C, an agar plug with a diameter of 4 mm was made at the end of the hyphae and subsequently inoculated on a V8A with each of five different fungicides. Each triplicated culture, inoculated with each isolate, was incubated in the dark at 25 °C, and mycelial growth was measured daily when the diameter of the untreated control group reached 60 mm. The percentage of mycelial growth inhibition for each isolate was measured in relation to the control. The concentrations required to inhibit 50% of the mycelial growth (EC50 values) were calculated as described previously [Citation53].
Table 2. Sensitivity ranges and mean values of effective concentrations to inhibit mycelial growth of Phytophthora nagaii and P. tentaculata isolates by EC50 values for five anti-oomycete fungicides, metalaxyl, ethaboxam, fluazinam, dimethomorph, and picarbutrazox.
3. Results and discussion
3.1. Cultural and morphological identification
The present study presented a detailed description of the cultural and morphological characteristics of four Korean isolates of Phytophthora (KACC 45737, 40909, 40912, and 40913) (). These fungal-like organisms exhibit filamentous, highly branched hyphae that are generally nonsegmented and contain multiple nuclei. The shape and size of sporangia differ among the isolates (), giving rise to mobile zoospores. Thick-walled oospores were produced in soil extract ().
Figure 1. Cultural and morphological characters of Phytophthora nagaii (A–C and G–H) and Phytophthora tentaculata (D–F and I–J). (A–C) Colonies of P. nagaii observed after 6 days of inoculation on potato dextrose agar (A), V8 agar (B), and cornmeal agar (C); (E–F) Colonies of P. tentaculata on potato dextrose agar (D), V8 agar (E), and cornmeal agar (F); (G) Subspherical sporangium of P. nagaii; (H) Oospore of P. nagaii; (I) Spherical, papillate sporangium of P. tentaculata; (J) Oogonium of P. tentaculata. Sources: KACC 45737 for P. nagaii and KACC 40912 for P. tentaculata.
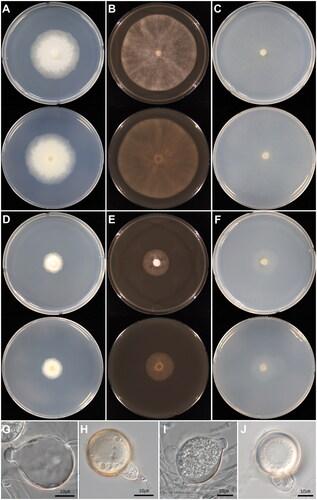
Colonies of the isolates KACC 45737 grew colorlessly in a radiate pattern with few aerial mycelia on three media, PDA, V8A, and CMA. The shape and features of the sporangia, sporangiophores, oogonia, oospores, and antheridia closely mirrored those typically found in Phytophthora nagaii [Citation47]. On host plant leaf cultures, this isolate mainly produces papillated sporangia (), although non-papillate forms were also noticed.
In the KACC 40909, 40912, and 40913, colonies exhibited a similar pattern as P. nagaii (KACC 45737) but displayed notable morphological distinctions. The colonies were smaller in diameter, and these isolates predominantly generated sporangia with a papilla when grown in soil extract, exhibiting diverse shapes. The morphological aspects of their sporangiophores, oogonia, and antheridia matched those of Phytophthora tentaculata [Citation54] but differed slightly in size and shape.
These detailed descriptions of P. nagaii (KACC 45737) and P. tentaculata (KACC 40909, 40912, and 40913) provide valuable insights into their morphology and growth characteristics. The information is crucial for accurately identifying and distinguishing these species, whether in the field or in a laboratory. These findings can enhance our understanding of Phytophthora biology and help in species differentiation and identification, potentially aiding in plant disease management and control.
3.2. Molecular phylogenetic identification
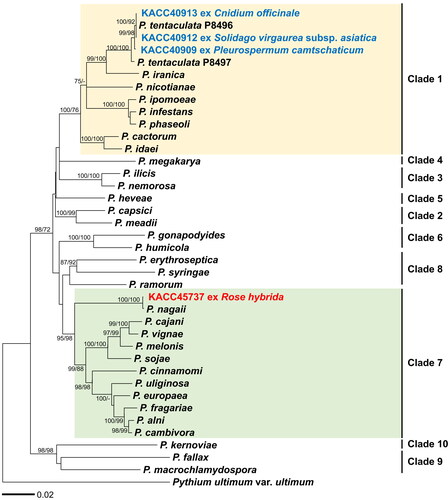
Sequence analysis was performed on the ribosomal ITS region and mitochondrial cox1 and cox2 gene sequences. Through a BLASTn search, KACC 40909 isolate showed high sequence similarities with the reference strain of Phytophthora tentaculata ex-type CPHST BL29 across all three markers: ITS sequence at 830/831 bp (MG865591; 99.88%), cox1 at 681/682 bp (MH136983; 99.85%), and cox2 at 476/478 bp (JF771611; 99.58%). Similarly, isolate KACC 40912 matched the ex-type of P. tentaculata (CPHST BL29) in ITS sequences at 830/831 bp (MG865591; 99.88%), cox1 at 694/694 bp (MH136983; 100%), and cox2 at 482/483 bp (JF771611; 99.79%). Also, KACC 40913 aligned with the identical reference of P. tentaculata in ITS at 828/829 bp (MG865591; 99.88%) but showed 100% identity for cox1 (MH136983) and cox2 (JF771611) sequences. Lastly, isolate KACC 45737 matched the same reference of P. tentaculata in the ITS at 854/855 bp (MG865547, 99.88%) and cox2 at 545/546 bp (LC596024, 99.82%), while showing a 100% identity in cox1 sequence (MH136940). Notably, KACC45737 was identical to Phytophthora nagaii in cox1 but exhibited a single nucleotide difference in ITS and cox2 sequences.
For the molecular phylogenetic identification of these four KACC isolates, representative species from clades 1–10 of the genus Phytophthora were selected from the World Phytophthora Collection (WPC). The sequences of three markers (ITS, cox1, cox2) were aligned to standardize all sequence lengths. This resulted in sequence lengths of 1002 bp for the ITS region, 654 bp for the cox1 gene, and 455 bp for the cox2 gene. Phylogenetic trees were constructed using both ML and ME methods based on the concatenated alignment of the three markers. In the phylogenetic tree (), KACC 40909, 40912, and 40913 formed a well-supported group with the reference sequences of P. tentaculata. Interestingly, the Korean isolates formed two subgroups by three nucleotide differences in cox1 and cox2 sequences. Meanwhile, KACC 45737 grouped with P. nagaii, demonstrating a high bootstrapping value of 100%.
Figure 2. Minimum evolution of Phytophthora species based on a concatenated alignment of the ITS rDNA, cox1, and cox2 mtDNA sequences. Bootstrapping values (minimum evolution/maximum likelihood) higher than 70% were shown above/below the branches (1000 replicates). A yellow box presents Clade 1, containing Phytophthora tentaculata, whereas a green box means Clade 7, containing Phytophthora nagaii.
3.3. Activities to anti-oomycete fungicides
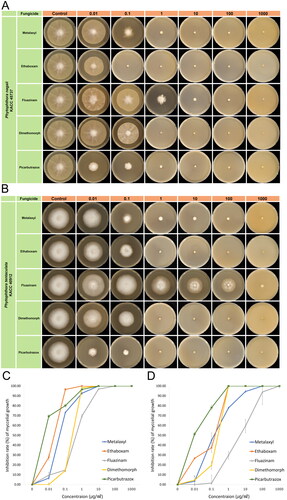
This study tested the sensitivity of two Phytophthora species against five different fungicides (metalaxyl, ethaboxam, dimethomorph, fluazinam, and picarbutrazox). Sensitivity was determined by observing if the colony growth on the fungicide media fell below an EC50 threshold of 1 μg/mL [Citation34]. For P. nagaii KACC45737, the EC50 value range (mean) for each fungicide was 0.02468 to 0.08478 μg/mL (0.05868) for metalaxyl, 0.01782 to 0.02356 μg/mL (0.02065) for ethaboxam, 0.42886 to 0.55275 μg/mL (0.48973) for fluazinam, 0.6478 to 0.7202 μg/mL (0.68215) for dimethomorph, and 0.00007 to 0.00021 μg/mL (0.00014) μg/mL for picarbutrazox (). As a result, the isolate displayed a high level of susceptibility to the five fungicides, with picarbutrazox showing the highest sensitivity.
For of P. tentaculata KACC40912, the EC50 value range (mean) was 0.16966 to 0.39068 (0.28566) μg/mL for metalaxyl, 0.06527 to 0.08145 (0.07452) μg/mL for ethaboxam, 4.74457 to 16.8783 (9.11459) μg/mL for fluazinam, 0.49248 to 0.63706 (0.57110) μg/mL for dimethomorph, and 0.00264 to 0.00536 (0.00375) μg/mL for picarbutrazox (). Based on these results, P. tentaculata isolate showed sensitivity to metalaxyl, ethaboxam, dimethomorph, and picarbutrazox but less sensitive to fluazinam.
The EC50 measurements for inhibiting the mycelial growth of P. nagaii and P. tentaculata with metalaxyl, ethaboxam, and dimethomorph were comparable to those observed for other Phytophthora species, such as P. agathidicida, P. cactorum, P. citrophthora, P. capsici, P. parasitica, and P. sojae [Citation55–59]. Both species were most sensitive to picarbutrazox among all the fungicides tested. Previously, fluazinam exhibited an EC50 value ranging from 0.14 to 0.27 against P. infestans [Citation60]. However, in the present study, P. tentaculata displayed high mycelial growth on fluazinam-containing media, compared to other Phytophthora species, suggesting its resistance to this particular fungicide, unlike other Phytophthora species.
The effects of varying fungicide concentrations on the growth of the mycelia of both species were visually depicted in , providing further insight into their reactions to different levels of fungicides. This study provided crucial information for understanding the sensitivity of two Phytophthora species to the most widely used anti-oomycete fungicides, which can guide targeted treatment strategies and help create more effective control methods. The distinct resistance of P. tentaculata to fluazinam compared to P. nagaii is of particular interest, suggesting species-specific responses that may require customized approaches in fungicide application. The notable sensitivity of both species to picarbutrazox underscores its promise as an effective agent for controlling these pathogens. Future research might focus on the mechanisms behind these sensitivity patterns and explore the field efficacy of these fungicides.
Figure 3. Mycelial growth of Phytophthora nagaii (A and C) and Phytophthora tentaculata (B and D) on V8 agar media, containing different concentrations of anti-oomycete fungicides (0, 0.01, 0.1, 1, 10, 100, and 1000 ug/mL). Agar plugs sourced from seven-days-old colonies were inoculated on V8A, with daily measurements of mycelial diameter until the control group reached a diameter of 60 mm.
4. Taxonomy
4.1. Phytophthora nagaii M.Z. Rahman, S. Uematsu, Toru Takeuchi, K. Shirai & Kageyama, Journal of General Plant Pathology 80(4): 353 (2014) [MB#804991]
4.1.1. Description
Colonies grow colorlessly with a radiate pattern and few aerial mycelia on PDA, V8A, and CMA at 25 °C, submerged growth on CMA, and measured 40–50 mm on PDA, 75–80 mm on V8A, and 75–85 mm in diameter on CMA after 72 h. The isolate produces mostly papillate but often non-papillate sporangia abundantly. Sporangia are terminal, single, ellipsoid often with tapering bases, and measured (25.2–) 26.6–32.7 (–34.4) × (18.4–) 20.2–24.9 (–26.2) (av. 29.6 × 22.5) um (n = 50). Sporangiophores are hyalin, simple sympodial and show eccentric basal attachment to sporangia. Oogonia are hyalin, mostly spherical and occasionally funnel-shaped with tapering bases and short stalks. Oospores are dark brown, aplerotic, mostly spherical, with a thick wall, and measured (35.2–) 36.1–39.1 (–40.1) (av. 37.6) um in diameter (n = 50). Antheridia are hyalin, predominantly paragynous and sometimes amphigynous.
4.1.2 Host plant
Rosa hybrida (Rosaceae)
4.1.3. Notes
Phytophthora nagaii isolates were previously classified under Phytophthora megasperma when discovered on roses in Japan [Citation61]. However, a later, in-depth phylogenetic analysis recognized it as a distinct species [Citation47]. While the initial documentation described this species as generating non-papillate sporangia when grown on grass blades [Citation47], our current research indicates that the species more frequently produces papillate than non-papillate sporangia. The recent discovery of P. nagaii in Korea could have wide-ranging implications, particularly in agriculture and plant disease management, given its impact on economically significant roses. This study contributes to our understanding, as its precise identification and morphological features may lead to improved treatment and control measures. The potential risk mandates ongoing investigation and monitoring.
4.2. Phytophthora tentaculata Kröber & Marwitz, Z. Pflanzenkrankh. Pflanzenschutz 100: 251 (1993) [MB#360186]
4.2.1. Description
Colonies grow colorlessly with a radiate pattern and few aerial mycelia on PDA, V8A, and CMA at 25 °C, submerged growth on CMA, and measured 15–20 mm in diameter on PDA and V8A, and 20–25 mm on CMA after 72 h. The isolates produce mostly papillate sporangia abundantly in soil extract. Sporangia are mostly ellipsoidal but often ovoid or elongated and measured (26.2-) 28.4–35.8 (-39) × (21.3-) 23.8–29.1 (-30.4) (av. 32.1 × 26.4) um (n = 50). Sporangiophores are hyalin and have branching points. Oogonia are hyalin and spherical to subglobose. Oospores are hyalin, aplerotic, spherical, and measured (23-) 25.7 − 33.3 (-36.9) (av. 29.5) um in diameter (n = 50). Antheridia are hyalin, predominantly amphigynous.
4.2.2. Host plant
Cnidium officinale (Apiaceae), Pleurospermum camtschaticum (Apiaceae), and Solidago virgaurea subsp. asiatica (Asteraceae)
4.2.3. Notes
This plant pathogen Phytophthora tentaculata, known for causing root and stalk rot, was initially isolated in a German nursery in 1993, where it affected plants such as Chrysanthemum, Verbena, and Delphinium species [Citation54]. Due to its ability to cause substantial economic harm to both the nursery industry and native plant species [Citation62], it is considered one of the top five most concerning Phytophthora species in the United States [Citation63,Citation64]. Later, this pathogen was also discovered in Spain, affecting Verbena plants [Citation65]. Recent studies have shown the host range of P. tentaculata to be broader, including Santolina chamaecyparissus (Lavender cotton) in Spain [Citation66], Origanum vulgare (Oregano) in Italy [Citation67], and Aucklandia lappa in China [Citation68]. This study isolated the Korean strains from Cnidium officinale (Apiaceae), Pleurospermum camtschaticum (Apiaceae), and Solidago virgaurea subsp. asiatica (Asteraceae). These findings support the broad host range of this pathogen and suggest that it can cause significant damage on various plants, not just the hosts recorded in this study. Considering its potential economic impact in Korea, there is an urgent need for further studies to explore its host range and financial impact within the country.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Abad ZG, Burgess TI, Redford AJ, et al. IDphy: an international online resource for molecular and morphological identification of Phytophthora. Plant Dis. 2023;107(4):987–998. doi: 10.1094/PDIS-02-22-0448-FE.
- Erwin DC, Ribeiro OK. Phytophthora diseases worldwide. St. Paul, Minnesota, USA: American Phytopathological Society; 1996.
- Kroon LP, Brouwer H, de Cock AW, et al. The genus Phytophthora anno 2012. Phytopathology. 2012;102(4):348–364. doi: 10.1094/PHYTO-01-11-0025.
- Balci Y, Bienapfl JC. Phytophthora in US forests. In: Lamour K, editor. Phytophthora: a global perspective. Boston: CABI Wallingford UK; 2013. p. 135–145.
- Parke JL, Knaus BJ, Fieland VJ, et al. Phytophthora community structure analyses in Oregon nurseries inform systems approaches to disease management. Phytopathology. 2014;104(10):1052–1062. doi: 10.1094/PHYTO-01-14-0014-R.
- Brasier C, Brown A. Infection of tree stems by zoospores of Phytophthora ramorum and P. kernoviae. In: Frankel SJK, Palmieri JT, Katharine M, editors. Proceedings of the sudden oak death third science symposium. Albany (CA): Department of Agriculture, Forest Service, Pacific Southwest Research Station; 2008. p. 167–168.
- Von Broembsen S, Brits G. Evaluation of the resistance of pincushion (Leucospermum spp.) breeding lines to root rot caused by Phytophthora cinnamomi. Acta Hortic. 1990;(264):115–122. doi: 10.17660/ActaHortic.1990.264.14.
- Goss EM, Larsen M, Chastagner GA, et al. Population genetic analysis infers migration pathways of Phytophthora ramorum in US nurseries. PLoS Pathog. 2009;5(9):e1000583. doi: 10.1371/journal.ppat.1000583.
- Hansen EM. Alien Forest pathogens: Phytophthora species are changing world forests. Boreal Environ Res. 2008;13:33–41.
- Hansen EM. Phytophthora species emerging as pathogens of Forest trees. Curr Forestry Rep. 2015;1(1):16–24. doi: 10.1007/s40725-015-0007-7.
- Rizzo D, Garbelotto M, Davidson J, et al. Phytophthora ramorum as the cause of extensive mortality of Quercus spp. and Lithocarpus densiflorus in California. Plant Dis. 2002;86(3):205–214. doi: 10.1094/PDIS.2002.86.3.205.
- Hyun I-H, Choi W. Phytophthora species, new threats to the plant health in Korea. Plant Pathol J. 2014;30(4):331–342. doi: 10.5423/PPJ.RW.07.2014.0068.
- KSPP. List of plant disease in Korea. Korea: Korean Society of Plant Pathology; 2022.
- Jee H, Kim W, Lee S, et al. Phytophthora cryptogea causing the foot rot of Gerbera jamesonii in Korea. Korean J Plant Pathol. 1996;12:374–376.
- Jee H-J, Kim W-G, Cho W-D. First report of Phytophthora palmivora isolated from areca palm and soil in Korea. Plant Pathol. J. 1997;13:438–441.
- Kyoung-Yul R. Stem rot of lily (Lilium L.) caused by Phytophthora cactorum in Korea. Korean J Plant Pathol. 1998;14:458–462.
- Jee H-J, Kim W-G, Cho W-D. Phytophthora root rot of Chinese cabbage and spinach caused by P. drechsleri in Korea. Plant Pathol J. 1999;15:28–33.
- Ryu K-Y, Kim J-S, Kim J-T, et al. First report of pink rot of potato (Solanum tuberosum) caused by Phytophthora erythroseptica in Korea. Res Plant Dis. 2003;9(1):32–35. doi: 10.5423/RPD.2003.9.1.032.
- Oh E, Lee S, Kim K, et al. First report of chestnut ink disease by Phytophthora katsurae on chestnut in Korea. Plant Dis. 2008;92(2):312–312. doi: 10.1094/PDIS-92-2-0312A.
- Lee J-K, Jo J-W, Shin K-C, et al. Isolation, identification and characterization of Phytophthora katsurae, causing chestnut ink disease in Korea. Plant Pathol J. 2009;25(2):121–127. doi: 10.5423/PPJ.2009.25.2.121.
- Kim B-S, Wai KPP, Siddique MI, et al. First report of Phytophthora leaf blight and vine rot of kudzu (Pueraria lobata) in Korea. Res Plant Dis. 2020;26(2):109–115. doi: 10.5423/RPD.2020.26.2.109.
- Jee H-J, Cho W-D, Kim W-G. Phytophthora diseases of apple in Korea: II. Occurrence of an unusual fruit rot caused by P. cactorum and P. cambivora. Plant Pathol J. 1997;13:145–151.
- Lee Y-H, Jee H-J, Cha K-H, et al. Occurrence of Phytophthora root rot on kiwifruit in Korea. Plant Pathol J. 2001;17:154–158.
- Jee H-J, Cho W-D, Kim C-H. Phytophthora diseases in Korea. Suwon, Korea: National Institute of Agricultural Science and Technology; 2000.
- Seo M-W, Song J-Y, Kim H-G. Multi-locus phylogeny analysis of Korean isolates of Phytophthora species based on sequence of ribosomal and mitochondrial DNA. Kor J Mycol. 2010;38(1):40–47. doi: 10.4489/KJM.2010.38.1.040.
- Nam B, Lee HJ, Choi Y-J. Organic farming allows balanced fungal and oomycetes communities. Microorganisms. 2023;11(5):1307. doi: 10.3390/microorganisms11051307.
- Nam B, Lee D-J, Choi Y-J. High-temperature-tolerant fungus and oomycetes in Korea, including Saksenaea longicolla sp. nov. Mycobiology. 2021;49(5):476–490. doi: 10.1080/12298093.2021.1985698.
- Han Y-K, Back C-G, Park M-J, et al. Control effects of fungicides against Fusarium wilt on watermelon and crown and foot rot on cucumber. KJPS. 2021;25(4):343–352. doi: 10.7585/kjps.2021.25.4.343.
- Jung S-K, Kim H-M, Ko J-A, et al. Chemical control of ivy stem rot disease. J Agric Life Sci. 2012;43:28–31.
- Kim B-S, Ahn J-W. Identification and fungicide responses of Phytophthora cactorum isolated from lily growing daekwallyong alpine area. Korean J Pest Sci. 2002;6:42–44.
- Kim D-S, Prak H-C, Chun S-J, et al. Field performance of a new fungicide ethaboxam against cucumber downy mildew, potato late blight and pepper Phytophthora blight in Korea. Plant Pathol J. 1999;15:48–52.
- Kim DS, Chun SJ, Jeon JJ, et al. Synthesis and fungicidal activity of ethaboxam against oomycetes. Pest Manag Sci. 2004;60(10):1007–1012. doi: 10.1002/ps.873.
- Kim J-S, Lee Y-G, Kwon M, et al. Mating types of Phytophthora infestans isolates and their responses to metalaxyl and dimethomorph in Korea. Res Plant Dis 2014;20(1):25–30. doi: 10.5423/RPD.2014.20.1.025.
- Lee S-M, Shin J-H, Kim S-B, et al. Characteristics of Phytophthora capsici causing pepper Phytophthora blight resistant to metalaxyl. Korean J Pest Sci. 2009;13:283–289.
- Shin J-H, Kim J-Y, Kim H-J, et al. Control efficacy of carboxylic acid amide fungicides against pepper Phytophthora blight causing Phytophthora capsici. Korean J Pest Sci. 2010;14:463–472.
- Zhang XZ, Ryu KY, Kim JS, et al. Changes in the sensitivity to metalaxyl, dimethomorph and ethaboxam of Phytophthora infestans in Korea. Plant Pathol J. 2005;21(1):33–38. doi: 10.5423/PPJ.2005.21.1.033.
- Tomlin C. The pesticide manual: a world compendium. United Kingdom: British Crop Protection Council; 2006.
- Davidse LC, Hofman AE, Velthuis GC. Specific interference of metalaxyl with endogenous RNA polymerase activity in isolated nuclei from Phytophthora megasperma f. sp. medicaginis. Exp Mycol. 1983;7(4):344–361. doi: 10.1016/0147-5975(83)90019-1.
- Uchida M, Roberson RW, Chun SJ, et al. In vivo effects of the fungicide ethaboxam on microtubule integrity in Phytophthora infestans. Pest Manag Sci. 2005;61(8):787–792. doi: 10.1002/ps.1045.
- Anema B, Bouwman J, Komyoji T, et al. Fluazinam: a novel fungicide for use against Phytophthora infestans in potatoes. Farnham (UK): British Crop Protection Council; 1992. p. 663–668.
- Komyoji T, Sugimoto K, Suzuki K. Effect of fluazinam, a new fungicide, on infection processes of several plant pathogenic fungi. Jpn J Phytopathol. 1995;61(2):145–149. doi: 10.3186/jjphytopath.61.145.
- Tucker R, Leaper D, Laidler S, editors. Fluazinam-control of potato late blight-UK experience 1992-3. Proceedings of the Brighton Crop Protection Conference – Pests and Diseases; Brighton, England; 1994.
- Davidse L, Looijen D, Turkensteen L. Van der Wal D. Occurrence of metalaxyl-resistant ttrains of Phytophthora infestans in Dutch potato fields. Eur J Plant Pathol. 1981;87:65–68.
- Dowley L, O'sullivan E. Metalaxyl-resistant strains of Phytophthora infestans (Mont.) de bary in Ireland. Potato Res. 1981;24(4):417–421. doi: 10.1007/BF02357324.
- Feng X, Baudoin A. First report of carboxylic acid amide fungicide resistance in Plasmopara viticola (grapevine downy mildew) in North America. Plant Health Prog. 2018;19(2):139–139. doi: 10.1094/PHP-01-18-0005-BR.
- Waterhouse G. Key to Pythium pringsheim. Commonwealth Mycol Inst Mycol Pap. 1967;109:1–15.
- Rahman MZ, Uematsu S, Takeuchi T, et al. Two new species, Phytophthora nagaii sp. nov. and P. fragariaefolia sp. nov., causing serious diseases on rose and strawberry plants, respectively, in Japan. J Gen Plant Pathol. 2014;80(4):348–365. doi: 10.1007/s10327-014-0519-1.
- White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York: Academic Press; 1990. p. 315–322.
- Robideau GP, De Cock AW, Coffey MD, et al. DNA barcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Mol Ecol Resour. 2011;11(6):1002–1011. doi: 10.1111/j.1755-0998.2011.03041.x.
- Hudspeth DS, Nadler SA, Hudspeth ME. A COX2 molecular phylogeny of the peronosporomycetes. Mycologia. 2000;92(4):674–684. doi: 10.1080/00275514.2000.12061208.
- Choi Y-J, Beakes G, Glockling S, et al. Towards a universal barcode of oomycetes–a comparison of the cox1 and cox2 loci. Mol Ecol Resour. 2015;15(6):1275–1288. doi: 10.1111/1755-0998.12398.
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010.
- Adaskaveg JE, Hao W, Förster H. Postharvest strategies for managing Phytophthora brown rot of citrus using potassium phosphite in combination with heat treatments. Plant Dis. 2015;99:1477–1482. doi: 10.1094/PDIS-01-15-0040-RE.
- Kröber H, Marwitz R. Phytophthora tentaculata sp. nov. and Phytophthora cinnamomi var. parvispora var. nov., zwei neue Pilze von Zierpflanzen in Deutschland. Z Pflanzenkr Pflanzenschutz. 1993;100:250–258.
- Matheron M, Porchas M. Impact of azoxystrobin, dimethomorph, fluazinam, fosetyl-Al, and metalaxyl on growth, sporulation, and zoospore cyst germination of three Phytophthora spp. Plant Dis. 2000;84(4):454–458. doi: 10.1094/PDIS.2000.84.4.454.
- Parra G, Ristaino JB. Resistance to mefenoxam and metalaxyl among field isolates of Phytophthora capsici causing Phytophthora blight of bell pepper. Plant Dis. 2001;85(10):1069–1075. doi: 10.1094/PDIS.2001.85.10.1069.
- Ali A, Kumar R, Mazákova J, et al. Evaluation of the ability of seven active ingredients of fungicides to suppress Phytophthora cactorum at diverse life stages, and variability in resistance found among isolates. JoF. 2022;8(10):1039. doi: 10.3390/jof8101039.
- McCoy AG, Noel ZA, Jacobs JL, et al. Phytophthora sojae pathotype distribution and fungicide sensitivity in Michigan. Plant Dis. 2022;106(2):425–431. doi: 10.1094/PDIS-03-21-0443-RE.
- Thurston AM, Waller L, Condron L, et al. Sensitivity of the soil-borne pathogen Phytophthora agathidicida, the causal agent of kauri dieback, to the anti-oomycete fungicides ethaboxam, fluopicolide, mandipropamid, and oxathiapiprolin. N Z Plant Prot. 2022;75:14–18.
- Rekanović E, Potočnik I, Milijašević-Marčić S, et al. Sensitivity of Phytophthora infestans (Mont.) de bary isolates to fluazinam, fosetyl-Al and propamocarb-hydrochloride. Pestic Fitomed. 2011;26:111–116.
- Nagai Y, Takeuchi T, Watanabe T. A stem blight of rose caused by Phytophthora megasperma. Phytopathology. 1978;68(5):684–688. doi: 10.1094/Phyto-68-684.
- Sims LL, Garbelotto M. Susceptibility to the rare Phytophthora tentaculata and to the widespread Phytophthora cactorum is consistent with host ecology and history. For Pathol. 2018;48:e12446.
- Sullivan M, Bulluck R. Phytophthora species in the environment and nursery settings. USA: United States Department of Agriculture; 2010.
- Rooney-Latham S, Blomquist C. First report of root and stem rot caused by Phytophthora tentaculata on Mimulus aurantiacus in North America. Plant Dis. 2014;98(7):996–996. doi: 10.1094/PDIS-09-13-1002-PDN.
- Moralejo E, Pérez‐Sierra AM, Álvarez L, et al. Multiple alien Phytophthora taxa discovered on diseased ornamental plants in Spain. Plant Pathol. 2009;58(1):100–110. doi: 10.1111/j.1365-3059.2008.01930.x.
- Álvarez L, Pérez-Sierra A, León M, et al. Lavender cotton root rot: a new host of Phytophthora tentaculata found in Spain. Plant Dis. 2006;90(4):523–523. doi: 10.1094/PD-90-0523A.
- Martini P, Pane A, Raudino F, et al. First report of Phytophthora tentaculata causing root and stem rot of oregano in Italy. Plant Dis. 2009;93(8):843–843. doi: 10.1094/PDIS-93-8-0843B.
- Meng J, Wang Y. First report of stalk rot caused by Phytophthora tentaculata on Aucklandia lappa in China. Plant Dis. 2008;92(9):1365–1365. doi: 10.1094/PDIS-92-9-1365B.
