Abstract
The genus Fuscoporia (Hymenochaetaceae, Basidiomycota) comprises poroid white-rot fungi characterized by dark brown hymenial setae, a dimitic hyphal system, and encrusted generative hyphae. Despite the ecological and commercial significance of Fuscoporia species, their identification has been challenging owing to their morphological overlap with other genera of Hymenochaetaceae and to the limited resolution of nuclear ribosomal DNA markers. With the advances in molecular research, Fuscoporia has been revised to include species from Inonotus sensu lato and Phellinus sensu lato, and 71 new species have been reported over the past decade. In Korea, a comprehensive taxonomic study elucidating the true diversity of Fuscoporia is yet to be conducted. Among the 11 Fuscoporia species reported in Korea, two were identified solely based on morphological characteristics, and four were identified based on nuclear ribosomal DNA regions, which have limited resolution for species identification in Fuscoporia. To investigate the current status of Fuscoporia species native to Korea, we conducted a phylogenetic study using four genetic markers (ITS + nrLSU + RPB2 + TEF1), along with morphological characteristics, and re-analyzed the GenBank records deposited from Korea. Ten Fuscoporia species were identified, including three previously unrecorded species. A detailed description of the unrecorded species and a list of proposed Korean names for all Fuscoporia species in Korea are provided. This study will guide further taxonomic and applied research of Fuscoporia by providing a species identification key and a verified multigenetic database, in addition to confirming the sequences in public database.
1. Introduction
The genus Fuscoporia Murrill in the family Hymenochaetaceae (Hymenochaetales, Basidiomycota) comprises yellowish-to-rusty poroid fungal species that cause white rot. They play essential roles in the forest ecosystem, acting as wood decomposers or parasites on diverse angiosperm and gymnosperm species [Citation1,Citation2]. Some Fuscoporia species, such as F. gilva (Schwein.) T. Wagner & M. Fisch and F. rhabarbarina (Berk.) Groposo, Log.-Leite & Góes-Neto, have commercial value for their known antibacterial, antiviral, and antioxidant properties [Citation3–6].
Fuscoporia, typified by Fuscoporia ferruginosa (Schrader) Murrill, was first proposed by Murrill in 1907 and later defined by morphological characteristics such as resupinate to pileate basidiomes, smooth, thin-basidiospores, dimitic hyphal system with stellate crystals at the end of the generative hyphae, and presence of brownish setae [Citation7]. However, owing to its morphological similarity to the other genera of Hymenochaetaceae, it was not widely accepted as an independent genus and was instead considered synonymous with Phellinus. Fuscoporia became widely accepted only when Wagner and Fischer [Citation8] reported the molecular phylogeny of Hymenochaetales based on nuclear large subunit (nrLSU) sequences, revising several genera and transferring many species to Fuscoporia. Subsequently, new Fuscoporia species have been reported worldwide based on multigenetic phylogeny [Citation9–15].
In the Republic of Korea, Fuscoporia species had been classified as Phellinus until two species, namely F. ferrea (Pers.) G. Cunn. and F. senex (Nees & Mont.) Ghob.-Nejh., were reported as Fuscoporia based on internal transcribed spacer (ITS) + nrLSU sequences, and four species, namely F. contigua (Pers.) G. Cunn., F. ferruginosa, F. gilva, and F. viticola (Schwein.) Murrill, were transferred from Phellinus [Citation16,Citation17]. Subsequently, the following five new species were reported based on multigenetic phylogeny and detailed morphological observations: F. dolichoseta Y. Cho, D. Kim & Y. W. Lim, F. gilvoides Y. Cho, D. Kim & Y. W. Lim, F. koreana Y. Cho, D. Kim & Y. W. Lim, F. reticulata Y. Cho, D. Kim & Y. W. Lim, and F. semicephala Y. Cho, D. Kim & Y. W. Lim [Citation18]. In summary, 11 Fuscoporia species have been reported in Korea. However, two Fuscoporia species, namely F. contigua and F. viticola, were identified based solely on the morphological descriptions of European or North American species without supporting molecular evidence. Furthermore, re-identification is necessary for the four Fuscoporia species previously identified through ITS + nrLSU: F. ferrea, F. ferruginosa, F. gilva, and F. senex [Citation16,Citation17], as recent studies have consistently reported the limited resolution of ribosomal DNA sequences within the genus Fuscoporia [Citation9,Citation18].
The main objective of the present study was to investigate the true diversity of Fuscoporia in Korea by applying the current taxonomic revisions. To achieve this, we conducted a phylogenetic analysis on unstudied Fuscoporia specimens and all GenBank sequences of Fuscoporia deposited from Korea using four genetic markers: ITS + nrLSU + RNA polymerase II subunit 2 gene (RPB2) + translation elongation factor 1 gene (TEF1). Our research shows that four of the 11 reported species are not phylogenetically verified to be present in Korea, while three unrecorded species are present, totaling 10 Fuscoporia species. We provide detailed morphological descriptions, sequences, a taxonomic key, and Korean names for all Fuscoporia species in Korea. This study contributes to the accurate identification of Fuscoporia species in Korea based on morphological and molecular analyses.
2. Materials and methods
2.1. Specimens studied
In total, 47 specimens were obtained from two fungaria in Korea: the Korea University Fungus Collection (KUC) and Seoul National University Fungus Collection (SFC) (Table S1). The specimens were collected from Korea between 2012 and 2023. Images and notes on the characteristics of fresh basidiomes were recorded in the collection field.
2.2. Molecular analysis
2.2.1. DNA extraction, PCR, and sequencing
Genomic DNA extraction was performed using 1 × 1 cm tissue samples from dried specimens with cetyltrimethylammonium bromide (CTAB) and the AccuPrep Genomic DNA Extraction Kit (Bioneer, Daejeon, South Korea) according to the manufacturer’s instructions.
Polymerase chain reaction (PCR) was conducted on a C1000 thermal cycler (Bio-Rad, Hercules, CA) using a PCR Premix (Bioneer, Daejeon, South Korea). The ITS region was amplified using the primer sets ITS1F/ITS4B [Citation19] with the following conditions: initial denaturation at 95 °C for 5 min; 35 cycles of 95 °C for 40 s, 55 °C for 40 s, and 72 °C for 60 s; and a final extension at 72 °C for 10 min. The nrLSU region was amplified using the primer sets LR0R/LR7 [Citation20] with the following conditions: initial denaturation at 95 °C for 5 min; 35 cycles of 95 °C for 40 s, 55 °C for 40 s, and 72 °C for 90 s; and a final extension at 72 °C for 10 min. For the protein-coding genes RPB2 and TEF1, the primer sets bRPB2-6F/bRPB2-7.1R [Citation21] and EF595F/EF1160R [Citation22] were used for PCR, respectively. The PCR conditions for RPB2 were as follows: initial denaturation at 94 °C for 2 min; 36 cycles of 94 °C for 45 s, 58 °C for 45 s, and 72 °C for 60 s; and a final extension at 72 °C for 10 min. The PCR conditions for TEF1 were as follows: initial denaturation at 95 °C for 4 min; 35 cycles of 95 °C for 30 s, 55 °C for 45 s, and 72 °C for 60 s; and a final extension at 72 °C for 10 min.
The PCR products were purified using the Expin™ PCR Purification Kit (GeneAll Biotechnology, Seoul, South Korea) and sequenced using an ABI 3730XL machine (Applied Biosystems, Waltham, MA) at Bioneer (Daejeon, South Korea) using PCR primer sets. The forward and backward sequences of each specimen were assembled using Geneious Prime 2023.0.4 (https://www.geneious.com). The final sequences have been deposited in GenBank ().
Table 1. List of Fuscoporia specimens and GenBank accessions used in the study.
2.2.2. Phylogenetic analysis
In addition to the 59 newly acquired sequences, 76 Fuscoporia sequences deposited from Korea were obtained from GenBank. For the phylogenetic analysis, a dataset of four genetic markers (ITS + nrLSU + RPB2 + TEF1) was constructed, including 201 published or type-derived sequences of Fuscoporia [Citation15,Citation18,Citation26]. Alignment, trimming, concatenation, model selection, and maximum-likelihood (ML) tree construction were performed using FunVIP ver. 0.3.23.1 (https://github.com/Changwanseo/FunVIP) with the “accurate” preset: alignment was performed by MAFFT ver. 7.453 [Citation33] and tree construction was performed by RAxML 8.2.12 [Citation34] with 1000 bootstrap replications. Three outgroup samples were used: Coniferiporia sulphurascens (FP-134848-SP), Coniferiporia weirii (FP-134801-SP), and Phellinidium asiaticum (Wei 5610) [Citation31,Citation32].
Subsequently, the newly acquired sequences and the GenBank sequences deposited from Korea were analyzed by a BLASTn search at the NCBI website against the nr/nt database (date accessed: August 10 2023), and ML trees were constructed based on two additional combinations (ITS and ITS + nrLSU) to confirm their identity and assess the accuracy of each method.
2.3. Morphological observation
Macroscopic characteristics such as color, length, width, and thickness of the pileus, context, and hymenophore were observed with the naked eye. The size of the pores and the margin of the hymenophore were observed using a Nikon SMZ1500 stereomicroscope (Nikon, Tokyo, Japan).
For the microscopic analysis, dried tissue from each specimen was sectioned and mounted in a drop of 5% KOH. The features of the hyphae, basidia, cystidia, basidiospores, and setae were observed under a Nikon 80i compound light microscope (Nikon, Tokyo, Japan) at ×400 magnification. A minimum of 20 elements were measured for each feature. For basidiospores, 5% of the extreme length and width values are provided in parentheses. “L”, “W”, and “Q” refer to the length, width, and length/width ratio of the basidiospores, respectively; “x/y” refers to the number of elements measured (x) and the number of specimens (y); “CB−” refers to acyanophilous in cotton blue; and “IKI−” refers to neither amyloid nor dextrinoid in Melzer’s reagent. All colors were described according to the “Methuen Handbook of Colour” [Citation35].
3. Results
Sequences of the four genetic markers were obtained from 17 Fuscoporia specimens (). The ITS + nrLSU + RPB2 + TEF1 dataset was constructed, comprising 345 sequences from 53 Fuscoporia species and three outgroup species. The concatenated sequence alignment contained 3218 bp, with ITS = 1–665, nrLSU = 666–2014, RPB2 = 2015–2689, and TEF1 = 2690–3218, including gaps. Fuscoporia species in Korea were re-identified by a BLASTn search and phylogenetic analysis based on three combinations of sequences (ITS, ITS + nrLSU, and ITS + nrLSU + RPB2 + TEF1) (Table S2). Two public Fuscoporia sequences (accession number: KA17-0255, accession number: KMCC04909) were excluded because they showed the highest similarity to Tapinella panuoides and Sanghuangporus vaninii based on a BLASTn search.
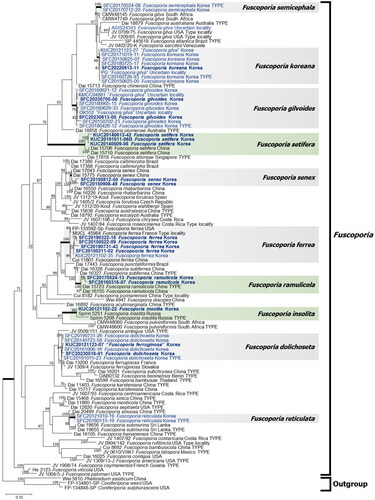
A total of 10 Korean Fuscoporia species were identified based on four genetic markers (). Eight out of the 10 clades were well-supported with high bootstrap values (>98). However, F. gilvoides had a moderate bootstrap value (75), and F. dolichoseta was not well-supported, having a low bootstrap value (<60). Seventeen newly analyzed specimens were divided into eight distinct taxa. Five of them corresponded to previously reported species, and three corresponded to unrecorded species in Korea. Seventy-six GenBank sequences submitted from Korea were identified as eight previously reported species in multigene phylogeny. However, in the BLASTn search, three species (F. dolichoseta, F. gilvoides, and F. senex) could not be identified because of conflicting top-hit results. These species also formed species complexes with other global Fuscoporia species in the ITS phylogeny. In the case of F. gilvoides, the ITS region was highly similar to that of F. karsteniana, whereas sequences of nrLSU, RPB2, and TEF1 regions were significantly different (Figure S1). In the ML tree based on the ITS + nrLSU region, F. senex formed a species complex with F. rhabarbarina.
Figure 1. Maximum-likelihood tree of Fuscoporia species based on four genetic markers (ITS + nrLSU + RPB2 + TEF1). Bootstrap values >60 are indicated at the nodes. A total of 10 species were identified, including three unrecorded species indicated by green color. Seven species indicated by gray color have been previously reported in Korea. Strains whose sequences were newly obtained in this study are indicated in bold, and sequences submitted from Korea are highlighted in blue.
Detailed descriptions of unrecorded species and species with limited morphological records are provided in the Taxonomy section. A list of Fuscoporia species in Korea with the newly proposed Korean names is also provided (Table S3).
4. Taxonomy
4.1. Unrecorded species in Korea
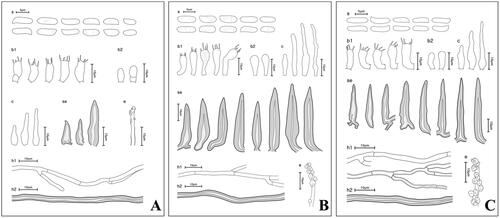
Fuscoporia insolita Spirin, Vlasák & Niemelä, Annales Botanici Fennici 51 (6): 404 (2014) ( and )
Description: Basidiomes perennial, resupinate, up to 2–6 mm thick. Pore surface nodulose, partly cracked, dark grayish brown (7F5); margin sterile, distinct, tomentose, light brown (6D8), fading to amber yellow (4B6), up to 2.5 mm wide. Pores circular to angular, grayish brown (8E3), 5–6 pores per mm; dissepiments entire. Tubes stratified, corky, grayish brown (7D3), up to 5 mm thick, each annual layer up to 2 mm thick. Subiculum corky, darker than tube layer, 1 mm thick.
Figure 2. Macroscopic morphology of unrecorded Fuscoporia species. (A) F. insolita (KUC20121102-22); (B) F. ramulicola (SFC20160316-07); (C) F. setifera (KUC20140509-06) (ba: basidiome; po: pore surface). Scale bar = 1 mm.
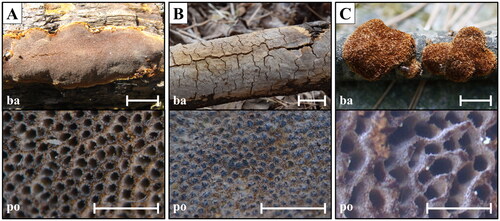
Figure 3. Microscopic morphology of unrecorded Fuscoporia species. (A) F. insolita; (B) F. ramulicola; (C) F. setifera (s: basidiospores; b1: basidia; b2: basidioles; c: cystidioles; se: setae; h1: generative hyphae; h2: skeletal hyphae; e: encrusted generative hyphae).
Hyphal system dimitic; generative hyphae thin-walled, branched, simple septate, hyaline, 2.0–2.7 μm wide in tubes, rare in subiculum, encrusted with crystals at dissepiment edge; skeletal hyphae dominant in both tubes and subiculum, thick-walled, unbranched, dark brown, 3.2–4.3 μm wide.
Basidia barrel-shaped or utriform with four sterigmata, simple septum at the base, 8.0–10.9(–11.6) × 4.1–5.1(–5.3) μm; basidioles smaller in size. Basidiospores cylindrical, thin-walled, smooth, some guttulate, hyaline, IKI−, CB−, (5.2–)5.4–7.6(–7.9) × 2.0–2.8 μm, L = 5.9 μm, W = 2.1 μm, Q = 2.7. Cystidioles conspicuous, narrowly lageniform, thin-walled, hyaline, 15.7–24.6 × 3.4–4.2 μm. Hymenial setae rare, obclavate or lanceolate, straight, acute at the apex, thick-walled, aseptate, dark brown, 14.2–26.1 × 3.8–6.0 μm. Mycelial setae absent.
Specimens examined: Korea, Gangwon-do, Pyeongchang-gun, Jinbu-myeon; 37°42′13″N 128°33′59″E, 872 m, Mt. Odae; November 2 2012 (KUC20121102-22).
Notes: Specimens from Korea have larger basidiospores than that of the holotype with (4.3–)4.7–7.2(–8.3) × (1.8–)1.9–2.4(–2.5) μm [Citation30].
Fuscoporia ramulicola (Pers.) Y.C. Dai & Q. Chen, MycoKeys 61: 82 (2019) ( and )
Description: Basidiomes annual, resupinate, inseparable, up to 1.5 mm thick. Pore surface cracked, grayish brown (6D3); margin sterile, paler than tubes, 1 mm wide. Pores circular to angular, 7–9 pores per mm; dissepiments entire. Tubes corky, orange grey (6B2), up to 1 mm deep. Subiculum corky, darker than tube layer, oak brown (5D6), up to 0.4 mm thick.
Hyphal system dimitic; generative hyphae thin-walled, branched, simple septate, hyaline, 1.8–3.0 μm wide in tubes, 1.7–2.6 μm wide in subiculum, encrusted with crystals at dissepiment edge; skeletal hyphae dominant in both tubes and subiculum, thick-walled, unbranched, rusty brown, 2.5–3.6 μm wide in tubes, 2.3–3.0 μm wide in subiculum.
Basidia clavate with four sterigmata, simple septum at the base, some with small guttules, 11.6–16.4 × 4.0–6.4 μm; basidioles smaller in size. Basidiospores cylindrical, thin-walled, smooth, mostly guttulate, hyaline, IKI−, CB−, (6.1–)6.3–7.7(–8.4) × (2.2–)2.4–3.0(–3.1) μm, L = 7.02 μm, W = 2.73 μm, Q = 2.59. Cystidioles fusoid, lageniform or cylindric-flexuous, thin-walled, hyaline, 10.9–39.7 × 2.5–4.3 μm. Hymenial setae subulate, acute at the apex, some are slightly bent at base, thick-walled, aseptate, dark brown, 14.2–44.4 × 3.7–8.7 μm. Mycelial setae absent.
Specimens examined: Korea, Gangwon-do, Yangyang-gun, Seo-myeon, Songeo-ri, 38°4′3.56″N 128°31′51.52″E, 240 m, on dead trunk of Alnus; March 16 2016 (SFC20160316-07). Korea, Jeollanam-do, Goheung-gun, Yeongnam-myeon, Paryeong-ro, Mt. Paryeong, on dead trunk of angiosperm; May 24 2017 (SFC20170524-13).
Notes: Specimens from Korea have larger basidia and basidiospores, and smaller hymenial setae than those of the holotype with basidia of 9–11 × 4.5–5.5 μm, basidiospores of (5.2–)5.8–7(–7.2) × (1.8–)2–2.5(–2.8) μm (L = 6.37 μm and W = 2.28 μm), and setae of 35–60 × 4.5–7 μm [Citation9]. Some larger cystidioles are found in the specimens from Korea, while the longest cystidiole recorded for the holotype from China is 22 μm [Citation9].
Fuscoporia setifera (T. Hatt.) Y.C. Dai, Fungal Diversity 45: 217 (2010) ( and )
≡ Phellinus setifer T. Hatt., Mycoscience 40(6): 483 (1999).
Description: Basidiomes annual, effused-reflexed, soft corky; pileus conchate, laterally fused, projecting up to 3 cm wide and up to 0.5 cm thick; pileal surface hairy, strigose, reddish brown (10F7), light reddish brown (6B6) near the margin. Pore surface paler than pileal surface, yellowish brown (5E6); margin distinct, hispid, 1–1.5 mm wide. Pores angular, 2–4 pores per mm; dissepiments entire at first, later dentate and protruding. Tubes corky, reddish brown (8D7), up to 2.5 mm deep. Context caramel (5C7), corky, thin.
Hyphal system dimitic; generative hyphae thin- to slightly thick-walled, branched, simple septate, hyaline, 2–3 μm wide in tubes and context, encrusted with crystals at dissepiment edge; skeletal hyphae dominant in both tubes and context, thick-walled, rarely branched, light brown, 2–4 μm wide in tubes and context.
Basidia clavate with four sterigmata, simple septum at the base, 10.0–17.1 × 3.9–6.9 μm; basidioles smaller in size. Basidiospores cylindrical, thin-walled, smooth, hyaline, IKI−, CB−, (5.7–)6.2–8.6(–8.9) × (1.9–)2.0–2.8(–3.0) μm, L = 7.45 μm, W = 2.37 μm, Q = 3.16. Cystidioles rare, cylindrical to narrowly lageniform, hyaline, thin-walled, 13.5–36.3 × 2.5–6.9 μm. Hymenial setae subulate, straight, acute at the apex, some with elongated base, thick-walled, aseptate, dark brown, 30.0–50.0 × 4.8–7.8 μm. Mycelial setae absent.
Specimens examined: Korea, Chungcheongbuk-do, Jecheon-si, Hansu-myeon, Songgye-ri 693-1; 36°49′13″N 128°6′0″E, 455 m, Mt. Worak; May 9 2014 (KUC20140509-06). Korea, Gyeongsangbuk-do, Cheongsong-gun, Juwangsan-myeon, Sangui-ri, Geumeungwang-i; 36°25′8″N 129°9′45″E, 789 m, Mt. Juwang; 11 Oct 2019 (KUC20191011-06B).
Notes: Specimens from Korea have longer basidiospores and larger cystidioles than those of the holotype specimen examined from Japan with basidiospores of 5.5–7.5 × 1.5–2.5 μm [Citation36].
4.2. Phylogenetically validated species in Korea
Fuscoporia ferrea (Pers.) G. Cunn., Bull. N.Z. Dept. Sci. Industr. Res. 73: 7 (1948) ( and )
Figure 4. Macroscopic morphology of Fuscoporia species. (A) F. ferrea (SFC20150211-02); (B) F. senex (SFC20150812-50). Scale bar = 1 mm.
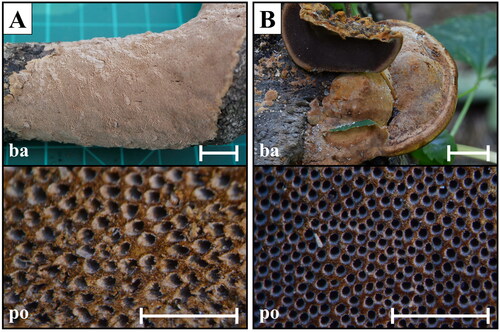
Figure 5. Microscopic morphology of Fuscoporia species. (A) F. ferrea; (B) F. senex (s: basidiospores; b1: basidia; b2: basidioles; c: cystidioles; se: setae; h1: generative hyphae; h2: skeletal hyphae; e: encrusted generative hyphae).
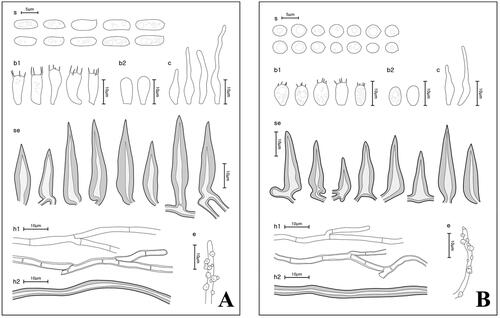
Description: Basidiomes perennial, resupinate, inseparable, up to 9 mm thick. Pore surface yellowish brown (5D8); margin sterile, paler than pore surface, 1–2 mm wide. Pores more or less circular, 5–6 pores per mm. Tubes corky, orange grey (6B2), up to 5 mm deep, dissepiments entire. Subiculum oak brown (5D6), corky, up to 4 mm thick.
Hyphal system dimitic; generative hyphae thin- to slightly thick-walled, branched, simple septate, hyaline to pale yellow, 2.1–2.9 μm wide in tubes, 1.8–2.7 μm wide in subiculum, encrusted with crystals at dissepiment edge; skeletal hyphae dominant in both tubes and subiculum, thick-walled, unbranched, rusty brown, 2.1–3.3 μm wide in tubes, 2.4–4.2 μm wide in subiculum.
Basidia clavate with four sterigmata, simple septum at the base, some with small guttules, 10.7–13.7 × 3.8–5.7 μm; basidioles smaller in size. Basidiospores cylindric, hyaline, thin-walled, smooth, mostly guttulate, IKI−, CB−, (5.4–)6.2–7.8(–8.2) × (2.3–)2.5–3.2(–3.3) μm, L = 7.03 μm, W = 2.80 μm, Q = 2.53. Cystidioles fusoid, lageniform or cylindric-flexuous, hyaline, thin-walled, 13.5–39.4 × 1.9–4.3 μm. Hymenial setae subulate, straight, acute at the apex, some with elongated base, dark brown, thick-walled, aseptate, 21.7–43.4 × 5.5–8.4 μm. Mycelial setae absent.
Specimens examined: Korea, Gangwon-do, Inje-gun, Girin-myeon, 37°55′5.12″N 128°24′15.83″E, 357 m, Mt. Bangtae Natural Recreational Forest; February 23 2015 (SFC20150211-02). Korea, Gangwon-do, Inje-gun, Girin-myeon, Jindong-ri, 38°9′11.77″N 128°23′23.61″E, Mt. Seorak; May 9 2015 (SFC20150522-09). Korea, Gyeongsangbuk-do, Bonghwa-gun, 37°4′23.25″N 128°57′39.77″E, Mt. Taebaek, Baekchoen valley, on dead trunk of angiosperm; March 22 2019 (SFC20190322-16) Korea, Gangwon-do, Taebaek-si; 37°6′34.30″N 128°56′43.17″E, Mt. Taebaek, on fallen branch of angiosperm; July 31 2019 (SFC20190731-43).
Notes: Korean specimens of Fuscoporia ferrea have slightly bigger basidiospores than holotype specimens (5–9 × 2–3 μm), but are identical in having perennial basidiomes and pore size of up to 6 per mm [Citation37]. This species has similar morphological characteristics with those of F. insolita and F. ramulicola in having cylindric basidiospores and no mycelial setae. Fuscoporia ferrea possessed abundant hymenial setae, whereas F. insolita has extremely rare hymenial setae [Citation30]. Also, F. ramulicola is different from F. ferrea in having smaller pore size (7–9 pores per mm) than those of F. ferrea.
Fuscoporia senex (Nees & Mont.) Ghob.-Nejh., in Ghobad-Nejhad & Dai, Mycotaxon 101: 208 (2007) ( and )
≡ Phellinus senex (Nees & Mont.) Imazeki, Bull. Govt Forest Exp. Stn Meguro 57: 115 (1952).
Description: Basidiomes perennial, pileate, solitary to imbricate, 2–10 cm wide, 0.3–2.1 cm thick. Pore surface chestnut brown (6F7); margin sterile, paler than pore surface, 1–2 mm wide. Pores circular, 7–10 pores per mm, dissepiments entire. Tubes corky, orange grey (6B2), up to 5 mm deep. Context tan (6E7), corky, up to 1.4 cm thick.
Hyphal system dimitic; generative hyphae thin- to thick-walled, branched, simple septate, hyaline, 1.8–2.5 μm wide in tubes, encrusted with crystals at dissepiment edge; skeletal hyphae dominant in both tubes and context, thick-walled, unbranched, rusty brown, 2.8–3.7 μm wide in tubes.
Basidia short clavate to barrel-shaped with four sterigmata, simple septum at the base, mostly guttulate, 7.1–10.7 × 4.0–5.6 μm; basidioles smaller in size. Basidiospores broadly ellipsoid to subglobose, thin-walled, smooth, mostly guttulate, hyaline, IKI−, CB−, (3.6–)3.8–4.5(–4.7) × (2.8–)3.0–3.7(–3.8) μm, L = 4.14 μm, W = 3.26 μm, Q = 1.28. Cystidioles rare, fusoid to lageniform, thin-walled, hyaline, 16.8–33.8 × 2.4–4.4 μm. Hymenial setae subulate, straight, acute at the apex, some with elongated base, thick-walled, aseptate, dark brown, 17.3–35.2 × 5.5–7.4 μm. Mycelial setae absent.
Specimens examined: Korea, Seoul, Jongno-gu, Jong-ro, Jongmyo; August 12 2014 (SFC20150812-50). Korea, Incheon, Ganghwa-gun, Hwado-myeon, Heungwang-ri; September 8 2016 (SFC20150908-49).
Notes: Korean specimens of F. senex have slightly smaller basidiospores compared to the specimens from Iran (3.6–4.5 × 2.5–3.2 μm) [Citation38]. Fuscoporia senex is different from other pileate Fuscoporia species in Korea in having smaller pore size (7–10 pores per mm).
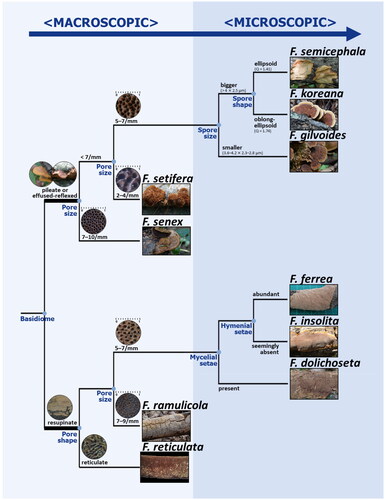
4.3. Taxonomic key to Fuscoporia species in Korea ()
1. Basidiomes effused-reflexed to pileate 2
1*. Basidiomes resupinate 6
2. Pores small, 7–10 per mm; Basidiospores broadly ellipsoid to subglobose, Q = 1.28 F. senex
2*. Pores 2–7 per mm 3
3. Pores big, 2–4 per mm F. setifera
3*. Pores 5–7 per mm 4
4. Basidiospores smaller, 3.6–4.1 × 2.3–2.8 μm, Q = 1.51 F. gilvoides
4*. Basidiospores longer, 3.9–4.8 μm long 5
5. Skeletal hyphae septate; Basidiospores 3.9–4.8 × 2.3–2.7 μm, Q = 1.74 F. koreana
5*. Skeletal hyphae aseptate; Basidiospores 4.0–4.8 × 2.8–3.4 μm, Q = 1.41 F. semicephala
6. Pores reticulate; mycelial setae present F. reticulata
6*. Pores circular to angular 7
7. Basidiospores ellipsoid to ovoid, 4.4–5.3 × 3.1–3.7 μm, Q = 1.44 F. dolichoseta
7*. Basidiospores cylindric 88. Hymenial setae extremely rare, seemingly absent F. insolita
8*. Hymenial setae abundant 9
9. Basidiomes thin, 1.5 mm, annual F. ramulicola
9*. Basidiomes thick, 9 mm, perennial F. ferrea
5. Discussion
In this study, 10 Fuscoporia species were verified through multigenetic marker phylogeny (), and their identification was further supported by morphological observations. Three of these (F. insolita, F. ramulicola, and F. setifera) were identified as species previously unrecorded in Korea. Descriptions are provided for three unrecorded species and two species that have not been previously described in detail. Additionally, errors in those previously identified only by ITS or ITS + nrLSU were confirmed by analyzing the GenBank sequences submitted from Korea. The sequences were derived from 24 Fuscoporia specimens: 21 from Korea and three from an uncertain locality.
The three previously unrecorded species reported in this study were originally described from Japan, China, and Far East Russia [Citation9,Citation30,Citation36], which are located relatively close to Korea. The specimens examined in this study showed minor variations in their morphological characteristics from the holotype description (see Notes for details). In particular, all three species had slightly larger basidiospores than those of the corresponding holotypes, which may be derived from the differences in measurement methods as well as regional variations. However, key characteristics other than basidiospores were consistent with the holotype. For example, specimens of F. insolita in Korea have seemingly absent, rare hymenial setae, which were regarded as the primary anatomic characteristics of F. insolita [Citation30]. Similarly, specimens of F. ramulicola exhibit annual, resupinate basidiomes with small pores and cylindrical basidiospores [Citation9], whereas F. setifera has a strigose pileus with larger pores and cylindrical basidiospores [Citation36]. These key characteristics are unique within the Fuscoporia species in Korea, making it relatively simple to identify the three unrecorded species.
The other seven Fuscoporia species verified in Korea are difficult to identify based solely on morphology, especially by macroscopic characteristics. For example, F. gilvoides, F. semicephala, and F. koreana exhibit a wide range of macromorphological features that overlap each other. However, their microscopic characteristics differed slightly; only F. koreana has septate skeletal hyphae, and F. gilvoides has smaller basidiospores [Citation18]. For correct identification in taxonomic and other applied studies, the verified specimens were carefully examined and a taxonomic key for Fuscoporia species in Korea was proposed (). Macroscopic characteristics were primarily used as criteria for the convenience of fieldwork, and microscopic characteristics were then used to identify species that were difficult to identify with macroscopic characteristics. Nevertheless, carrying out a phylogenetic analysis is recommended to distinguish all Fuscoporia species because their morphological differences are challenging to recognize.
Figure 6. Taxonomic key for 10 Korean Fuscoporia species. The branch length does not indicate the genetic distance between species. Four recorded species not supported with sequence evidence (F. contigua, F. ferruginosa, F. gilva, and F. viticola) were excluded.
The specimen previously reported as F. ferruginosa (KUC20121123-07) was re-identified as F. dolichoseta, and all specimens previously reported as F. gilva (KMCC04881, OK552, and KUC20121123-27) were re-identified as either F. gilvoides or F. koreana. F. ferruginosa is phylogenetically close to F. dolichoseta, forming a species complex in the ITS tree because of limited resolution. It also has a resupinate basidiome and mycelial setae, which can be easily confused with F. dolichoseta [Citation18,Citation39]. Likewise, F. gilva is a well-known species with typical brownish effused-reflexed to pileate basidiomes, which are extremely similar to those of numerous Fuscoporia species, including F. gilvoides and F. koreana. Therefore, many misidentified sequences of F. gilva have been submitted to GenBank without any type-derived sequences for correct identification [Citation18]. This makes identification challenging, especially using a BLASTn search. Fuscoporia gilva has been frequently analyzed in applied research owing to its anti-inflammatory and antioxidative properties [Citation3–5,Citation40]. Therefore, accurate identification is especially crucial for F. gilva to avoid the accumulation of erroneous data.
Additionally, four previously reported Fuscoporia species (F. contigua, F. ferruginosa, F. gilva, and F. viticola) were not verified to be present in Korea. The only F. gilva sequence uploaded from Korea (ASIS24343) was unpublished and had an uncertain locality. Considering that these species were initially reported from North America and New Zealand [Citation41,Citation42], are mainly found in North America, Europe, and Southwest China [Citation24,Citation26], and are morphologically similar to the Asian species reported in Korea, there is a high possibility that previous reports on these four species from Korea may have misidentified them. However, the limited sample size in this study cannot be overlooked, particularly given that Fuscoporia species are relatively frequent across the country. Therefore, collecting more specimens and verifying them through phylogenetic analysis is necessary to validate their presence in Korea.
In this study, eight out of 10 Fuscoporia species in Korea formed a distinct clade that was well supported in the ML tree constructed using a concatenated sequence dataset of ITS + nrLSU + RPB2 + TEF1. However, two species (F. dolichoseta and F. gilvoides) were not supported with high bootstrap values. As we included all records of Fuscoporia deposited from Korea, strains with only the ITS sequence were contained in the dataset. Also, we collected mainly the type-derived or type locality sequences for each species to exclude a few sequences that were suspected to contain errors, such as paraphyletic clade and swapping of sequences among species. This missing data and the use of fewer reference sequences may have lowered the bootstrap values compared to the previous multigene phylogenetic tree of Fuscoporia [Citation18]. Nevertheless, including additional genes for the phylogenetic tree can enhance the accuracy of analysis even when the gene is missing for some taxa [Citation43].
Our results of the ML tree inferred based only on the ITS region also addressed the importance of the multigene phylogenetic tree. Three clades formed species complexes in the ML tree inferred based only on the ITS region: (i) F. dolichoseta complex with four other species, (ii) F. gilvoides complex with F. gilva and F. karsteniana, and (iii) F. senex complex with F. rhabarbarina. Our result supports that assessing multigenetic markers in phylogenetic analyses increases reliability and resolution [Citation44]. This further indicates that the ITS region, as the genetic region commonly used for fungal identification, does not provide sufficient resolution within Fuscoporia. Therefore, analysis of multigenetic markers, including protein-coding genes, is necessary to accurately identify Fuscoporia species at the species level.
In conclusion, similarities in morphology and ITS sequences combined with misleading sequences in the public database might have resulted in previous misidentification of Fuscoporia in Korea. To address this problem, the present study provides a taxonomic key for morphological identification as well as a verified multigenetic database for phylogenetic analysis. This research will serve as the basis to accurately identify native Fuscoporia species and investigate their pharmaceutical properties.
Table S2.xlsx
Download MS Excel (22.1 KB)Table S1.xlsx
Download MS Excel (19.8 KB)Table S3.xlsx
Download MS Excel (17.4 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Karadelev M, Rusevska K, Stojanovskа S. Ecology and distribution of genus Phellinus (Hymenochaetaceae) in the Republic of Macedonia. Proceedings of the III Congress of Ecologists of the Republic of Macedonia with International Participation; Struga. Special issues of Macedonian Ecological Society 8; 2007. p. 197–207.
- Luana G, Fabiano S, Fabio G, et al. Comparing visual inspection of trees and molecular analysis of internal wood tissues for the diagnosis of wood decay fungi. Forestry. 2015;88(4):465–470. doi: 10.1093/forestry/cpv015.
- Chang ZQ, Gebru E, Lee SP, et al. In vitro antioxidant and anti-inflammatory activities of protocatechualdehyde isolated from Phellinus gilvus. J Nutr Sci Vitaminol. 2011;57(1):118–122. doi: 10.3177/jnsv.57.118.
- Duong TH, Dang NQ. Total phenolic, flavonoid content and antioxidative, α-amylase inhibitory activity of Phellinus gilvus fruiting body extracts. VNU J Sci Nat Sci Technol. 2022;38(1):71–185. doi: 10.25073/2588-1140/vnunst.5171.
- Sun Y, Zhong S, Deng B, et al. Impact of Phellinus gilvus mycelia on growth, immunity and fecal microbiota in weaned piglets. PeerJ. 2020;8:e9067. doi: 10.7717/peerj.9067.
- Walder R, Kalvatchev Z, Garzaro D, et al. Natural products from the tropical rain forest of Venezuela as inhibitors of HIV-1 replication. Acta Cient Venez. 1995;46(2):110–114.
- Fiasson JL, Niemelä T. The Hymenochaetales: a revision of the European poroid taxa. Karstenia. 1984;24(1):14–28. doi: 10.29203/ka.1984.224.
- Wagner T, Fischer M. Proceedings towards a natural classification of the worldwide taxa Phellinus s.l. and Inonotus s.l., and phylogenetic relationships of allied genera. Mycologia. 2002;94(6):998–1016.
- Chen Q, Dai YC. Two new species of Fuscoporia (Hymenochaetales, Basidiomycota) from southern China based on morphological characters and molecular evidence. Mycokeys. 2019;61:75–89. doi: 10.3897/mycokeys.61.46799.
- Chen Q, Liu L, Zhang DS, et al. Fuscoporia hainanensis sp. nov. (Hymenochaetales, Basidiomycota), a new member of the F. contigua group. Phytotaxa. 2022;558(3):251–262. doi: 10.11646/phytotaxa.558.3.1.
- Chen Q, Liu L, Si J, et al. Taxonomic and phylogenetic contributions to Fuscoporia (Hymenochaetales, Basidiomycota): two new species from Hawaii with a key to North American species. Front Cell Infect Microbiol. 2023;13:1205669. doi: 10.3389/fcimb.2023.1205669.
- Hussain S, Al-Kharousi M, Al-Muharabi MA, et al. Phylogeny, distribution and time divergence of Fuscoporia (Hymenochaetaceae, Basidiomycota) with the description of a new species from Dhofar region, southern part of Oman. Phytotaxa. 2022;570(2):150–164. doi: 10.11646/phytotaxa.570.2.3.
- Olou BA, Langer E, Ryvarden L, et al. New records and barcode sequence data of wood-inhabiting polypores in Benin with notes on their phylogenetic placements and distribution. Fungal Syst Evol. 2023;11(1):11–42. doi: 10.3114/fuse.2023.11.02.
- Raymundo T. Fuscoporia valenzuelae (Hymenochaetaceae, Basidiomycota), a new species from the tropical dry forest in Mexico. Acta Bot Mex. 2021;128:e1844.
- Wu F, Zhou LW, Vlasák J, et al. Global diversity and systematics of Hymenochaetaceae with poroid hymenophore. Fungal Divers. 2022;113(1):1–192. doi: 10.1007/s13225-021-00496-4.
- Jang Y, Lee SW, Jang S, et al. Four unrecorded wood decay fungi from Seoul in Korea. Mycobiology. 2012;40(3):195–201. doi: 10.5941/MYCO.2012.40.3.195.
- Jang Y, Jang S, Lee J, et al. Diversity of wood-inhabiting polyporoid and corticioid fungi in Odaesan National Park, Korea. Mycobiology. 2016;44(4):217–236. doi: 10.5941/MYCO.2016.44.4.217.
- Cho Y, Kim D, Lee Y, et al. Validation of Fuscoporia (Hymenochaetales, Basidiomycota) ITS sequences and five new species based on multi-marker phylogenetic and morphological analyses. IMA Fungus. 2023;14(1):12. doi: 10.1186/s43008-023-00117-6.
- Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2(2):113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x.
- Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172(8):4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990.
- Matheny PB. Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe; Agaricales). Mol Phylogenet Evol. 2005;35(1):1–20. doi: 10.1016/j.ympev.2004.11.014.
- Kauserud H, Schumacher T. Outcrossing or inbreeding: DNA markers provide evidence for type of reproductive mode in Phellinus nigrolimitatus (Basidiomycota). Mycol Res. 2001;105(6):676–683. doi: 10.1017/S0953756201004191.
- Du P, Chen Q, Vlasák J. Fuscoporia ambigua sp. nov., a new species from America and China. Phytotaxa. 2020;456(2):175–185. doi: 10.11646/phytotaxa.456.2.5.
- Chen Q, Wu F, Ji X-H, et al. Phylogeny of the genus Fuscoporia and taxonomic assessment of the F. contigua group. Mycologia. 2019;111(3):423–444. doi: 10.1080/00275514.2019.1570749.
- Pires RM, Motato-Vásquez V, de Gugliotta AM. Fuscoporia atlantica sp. nov., a new polypore from the Brazilian Atlantic Rainforest. Mycotaxon. 2015;130(3):843–855. doi: 10.5248/130.843.
- Chen Q, Du P, Vlasák J, et al. Global diversity and phylogeny of Fuscoporia (Hymenochaetales, Basidiomycota). Mycosphere. 2020;11(1):1477–1513. doi: 10.5943/mycosphere/11/1/10.
- Vlasák J, Kout J, Chan Q, et al. Fuscoporia caymanensis sp. nov. (Basidiomycota, Hymenochaetaceae), a new species from tropical America. Phytotaxa. 2020;472(2):1179–3155.
- Brazee NJ. Phylogenetic relationships among species of Phellinus sensu stricto, cause of white trunk rot of hardwoods, from Northern North America. Forests. 2015;6(11):4191–4211. doi: 10.3390/f6114191.
- Tchoumi JMT, Coetzee MPA, Rajchenberg M, et al. Poroid Hymenochaetaceae associated with trees showing wood-rot symptoms in the Garden Route National Park of South Africa. Mycologia. 2020;112(4):722–741. doi: 10.1080/00275514.2020.1753160.
- Spirin V, Vlasák J, Niemelä T. Fuscoporia insolita (Hymenochaetales, Basidiomycota), a new species from Russian Far East. Ann Bot Fenn. 2014;51(6):403–406. doi: 10.5735/085.051.0607.
- Zhou LW, Vlasák J, Dai YC. Taxonomy and phylogeny of Phellinidium (Hymenochaetales, Basidiomycota): a redefinition and the segregation of Coniferiporia gen. nov. for forest pathogens. Fungal Biol. 2016;120(8):988–1001. doi: 10.1016/j.funbio.2016.04.008.
- Wang XW, Liu SW, Zhou LW. An updated taxonomic framework of Hymenochaetales (Agaricomycetes, Basidiomycota). Mycosphere. 2023;14(1):452–496. doi: 10.5943/mycosphere/14/1/6.
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010.
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033.
- Kornerup A, Wanscher JH. Methuen handbook of colour. 3rd ed. London: Eyre Methuen; 1967.
- Hattori T. Phellinus setifer sp. nov. and P. acontextus, two noteworthy polypores from temperate areas of Japan, with notes on their allies. Mycoscience. 1999;40(6):483–490. doi: 10.1007/BF02461025.
- Bourdot H, Galzin A. Hyménomycètes de France (XI. Porés). Bull Soc Mycol France. 1925;41:247.
- Ghobad-Nejhad M, Dai YC. The genus Phellinus s.l. (Basidiomycota) in Iran. Mycotaxon. 2007;101:201–222.
- Núñez M, Ryvarden L. East Asian polypores 1. Ganodermataceae and Hymenochaetaceae. Synop Fung. 2000;13:1–168.
- Yoon KN, Jang H. Antioxidant and antimicrobial activities of fruiting bodies of Phellinus gilvus collected in Korea. Korean J Clin Lab Sci. 2016;48(4):355–364. doi: 10.15324/kjcls.2016.48.4.355.
- Cunningham GH. New Zealand Polyporaceae. 2. The genus Fuscoporia. Bull N Z Dept Ind Res. 1948;73:1–14.
- Murrill WA. Polyporaceae 1. North Am Flora. 1907;9:1–72.
- Jiang W, Chen S-Y, Wang H, et al. Should genes with missing data be excluded from phylogenetic analyses? Mol Phylogenet Evol. 2014;80:308–318. doi: 10.1016/j.ympev.2014.08.006.
- Sung GH, Sung JM, Hywel-Jones NL, et al. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): identification of localized incongruence using a combinational bootstrap approach. Mol Phylogenet Evol. 2007;44(3):1204–1223. doi: 10.1016/j.ympev.2007.03.011.
- Yuan H-S, Lu X, Dai Y-C, et al. Fungal diversity notes 1277–1386: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2020;104(1):1–266. doi: 10.1007/s13225-020-00461-7.
