Abstract
Golovinomyces (Erysiphaceae, Ascomycota) is an obligate plant pathogenic group causing powdery mildew on diverse angiosperm plants, including economically significant crops. Despite advancements in the taxonomy and phylogeny of Golovinomyces species using ribosomal DNA markers (ITS and LSU), several taxonomic issues remain unresolved. Previously, the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene, which exhibits higher nucleotide variation, has been proposed as an additional marker for powdery mildew species. In this study, we designed a new primer set (GoGPD-F and GoGPD-R) to improve the PCR success and efficiency of the GAPDH gene across various Golovinomyces species and dried herbarium specimens. The primers were successful in amplifying and sequencing the GAPDH gene in sixteen Golovinomyces species, including six species not previously registered in GenBank and two undescribed species. This development is a significant contribution to future research on the identification, taxonomy, and phylogeny of Golovinomyces species, offering a more robust tool for resolving existing taxonomic issues.
Powdery mildew fungi (Erysiphales; Ascomycota) are obligate plant pathogens responsible for significant economic and ecological impacts on various angiosperm plants worldwide. These fungi are characterized by white conidia and mycelia on the surfaces of leaves, stems, and flowers of their host plants. The genus Golovinomyces was initially classified under Erysiphe (e.g. E. cichoracearum) and later as Erysiphe section Golovinomyces [Citation1], but subsequent phylogenetic studies elevated it to the genus level [Citation2,Citation3]. This genus includes several notorious species, such as G. cichoracearum (syn. Erysiphe cichoracearum) parasitic on multiple plant families, G. bolayi on lettuce [Citation4], G. latisporus on sunflower [Citation5], and G. tabaci on cucurbits [Citation4].
Molecular phylogenetic approaches, mainly using ribosomal internal transcribed spacer (ITS) and large subunit (LSU) sequences, have advanced the understanding of taxonomic and phylogenetic relationships among Golovinomyces species [Citation4–8]. However, these markers often have limited informative sites, making it challenging to differentiate closely related species [Citation7] and providing insufficient resolution for species complexes within Golovinomyces, such as G. ambrosiae [Citation5], G. spadiceus [Citation5], and G. bolayi [Citation4]. Accurate identification of these pathogens is essential for effective disease control and management, necessitating the exploration of alternative genetic markers with higher variability and discriminatory power for this fungal group.
A multi-locus approach has recently been adopted in phylogenetic studies of powdery mildews [Citation5,Citation6,Citation9]. The rDNA intergenic spacer (IGS) region and the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene, in addition to ITS and LSU, have been proposed as supplementary phylogenetic markers [Citation6,Citation9]. Among these, the GAPDH gene is particularly effective in distinguishing closely related species due to its higher nucleotide polymorphisms. This gene has been widely used in the taxonomic and phylogenetic studies of various fungal groups [Citation10–12]. Despite its potential, previously designed primers showed low amplification success across Golovinomyces species and for dried herbarium specimens, limiting their effectiveness. The present study aimed to design a novel primer set improve their utility in taxonomic and phylogenetic studies.
Powdery mildew samples were collected from diverse host plants in Korea and deposited at the Korea University Herbarium (KUS-F). Genomic DNA was extracted from dried herbarium specimens of Golovinomyces and allied genera () using the MagListo 5 M Plant Genomic DNA Extraction Kit (Bioneer, Daejeon, Korea). To identify the samples, all were amplified with powdery mildew-specific primer sets PM10/ITS4 for ITS and PM3/TW14 for LSU rDNA regions [Citation2,Citation13,Citation14]. The PCR products were purified and bidirectionally sequenced by Macrogen (Seoul, Korea) with the same primers used for amplification.
Table 1. List of powdery mildew samples collected in Korea.
GAPDH gene sequences from twenty Golovinomyces species available in GenBank were aligned using SeqMan in the DNASTAR software package (Lasergen, Madison, WI, USA). A suitably variable region of ⁓323 bp was selected to design new primers, targeting conserved regions flanking the variable sites. Fourteen candidates for the forward primer were designed by modifying the existing primer PMGAPDH1 (GGAATGGCTATGCGTGTACC) [Citation9]. The protein codon site of PMGAPDH1 was confirmed using ExPASy (SIB Swiss Institute of Bioinformatics, Lausanne, Switzerland), and the third nucleotides of each codon, where mutations frequently occur [Citation15], were hybridized to the transition nucleotide. Four reverse primer candidates were designed in regions with low variation among the reference sequences, with lengths between 18 and 22 base pairs. For primer selection, the accuracy and efficiency of the candidate forward and reverse primers were compared through PCR tests and sequencing analysis. As a result, a primer set GoGPD-F (GGAATGGCYATGCGTGTRCC) and GoGPD-R (CAARGARATTCCRGCYTTTGC) was selected for its highest amplification rate and best sequence quality, yielding specific fragments ranging from 219 to 231 bp (excluding primers) of the GAPDH gene ().
Figure 1. Primer design on GAPDH sequences of Golovinomyces species. The newly designed primers (GoGPD-F and GoGPD-R) are shown above and below blue bars, while the previously used primers (PMGAPDH1 and PMGAPDH3R) are shown above and below red bars.

A gradient PCR was performed to determine the optimal amplification condition, identifying 55.1 °C as the ideal annealing temperature. The touchdown method, which decreases the annealing temperature by 0.2 °C per cycle, further increased amplification efficiency. The final PCR conditions were as follows: initial denaturation at 95 °C for 3 min, followed by 40 cycles of denaturation at 95 °C for 30 s, annealing at 55.1 °C (-0.2 °C per cycle) for 40 s, elongation at 72 °C for 40 s, and final elongation at 72 °C for 10 min. The reproducibility of this PCR method was verified using three commercial PCR mixtures: PerfectShot™ Ex Taq (Loading dye mix) (Takara Bio, Shiga, Japan), TOPsimple™ PCR DryMIX-nTaq (Enzynomics, Seoul, Republic of Korea), and AccuPrep® PCR/Gel Purification Kit (Bioneer, Daejeon, Korea). Additionally, all experiments were replicated using two different thermal cyclers in two separate laboratories: the Bio-Rad T100 Thermal Cycler (Bio-Rad Laboratories, Hercules, CA, USA) at Kunsan National University and the Thermo Fisher SimpliAmp™ Thermal Cycler (Thermo Fisher Scientific, Waltham, MA, USA) at Jeonbuk National University.
DNA samples from twenty-one Golovinomyces species () were PCR-amplified and Sanger-sequenced to compare the efficiency of the newly designed primers (GoGPD-F and GoGPD-R) and previously reported primers (PMGAPDH1 and PMGAPDH3R) [Citation9]. The new primers showed a higher species coverage (76%; sixteen Golovinomyces species) compared to the previously reported primers (42%; nine species). They also successfully sequenced six Golovinomyces species that had not been previously sequenced: G. adenophorae, G. arabidis, G. artermisiae, G. bolayi, G. macrocarpus, and G. tabaci. Additionally, the primers amplified two Erysiphe species (E. quercicola and E. sp.) but failed to amplify DNA from other genera such as Arthrocladiella, Blumeria, Cystotheca, Phyllactinia, Pleochata, and Sawadaea. The primers were effective in amplifying DNA from dried herbarium samples collected between 2006 and 2021, demonstrating their ability to amplify DNA from samples up to 18 years old. This broad amplification success underscores the applicability and effectiveness of the new primers for Golovinomyces species.
For the sixteen Golovinomyces species, the variability and divergence of the GAPDH sequences obtained using the newly developed primers were compared with those of ITS and LSU markers. The GAPDH exhibited a higher number of variable and informative sites (38% and 27%, respectively) compared to ITS (15% and 9%) and LSU (5% and 3%) regions (). Using the Kimura 2-parameter model, inter-species genetic divergence of the target gene ranged from 0.9% to 31.3%, which is higher than those of ITS (0–8.3%) and LSU (0–3.6%) markers. This comparative analysis indicates that GAPDH sequences provide higher resolution for species discrimination, effectively addressing the limitations of ITS and LSU markers.
Table 2. Sequence analysis of Golovinomyces species using the newly developed GAPDH primers and the ribosomal ITS and LSU primers.
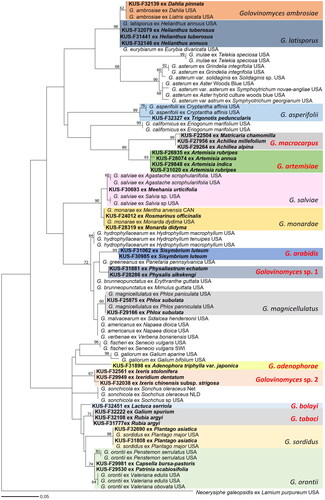
A minimum evolution tree was reconstructed using the newly obtained GAPDH sequences in MEGA 11 software [Citation16]. The confidence levels of each branch were evaluated using 1000 bootstrap replications. The GAPDH tree accurately grouped all analyzed samples into their respective species (), demonstrating its high effectiveness in differentiating Golovinomyces species. In the phylogenetic tree, the six species (G. adenophorae, G. arabidis, G. artermisiae, G. bolayi, G. macrocarpus, and G. tabaci) for which GAPDH sequences were obtained for the first time in this study formed distinct and unique groups, clearly separated from other known species. Additionally, two new species candidates of Golovinomyces were discovered; one parasitic on Physalis and Physaliastrum spp. and the other on Ixeris and Ixeridium spp. Notably, several species, including G. asperifolii, G. salviae, and G. orontii, formed subgroups, associated with specific host plant. Given the high host specificity of powdery mildew fungi, these subgroupings suggest the potential to classify them as separate species. For instance, G. ambrosiae s. lat., previously considered a single species based on rDNA sequences, was successfully distinguished by the GAPDH sequences it into two species, G. ambrosiae s. str. and G. latisporus, aligning with the findings of Qiu et al. [Citation5] and Bradshaw et al. [Citation8]. These findings highlight the efficacy of the GAPDH marker in resolving taxonomic issues of Golovinomyces species.
Figure 2. Maximum likelihood tree of Golovinomyces species based on GAPDH sequences. Bootstrapping support values higher than 60% are given at the branches. Korean specimens sequenced in this study are in bold. Golovinomyces species initially sequenced for the GAPDH gene in this study are highlighted in red. The scale bar equals the number of nucleotide substitution sites.
The present study successfully designed a novel primer set for the GAPDH gene, demonstrating a high success rate in amplifying, sequencing, and differentiating a broad range of Golovinomyces species. Therefore, the primers could serve as an effective alternative for future research on the taxonomy and phylogeny of Golovinomyces species.
Acknowledgement
The authors would like to express our gratitude to Prof. Hyeon-Dong Shin (Korea University) for collecting, identifying, and providing the powdery mildew specimens used in this study.
References
- Braun U. Beitrag zur systematik und nomenklatur der Erysiphales. Feddes Repertorium. 1978;88(9–10):655–665. doi: 10.1002/fedr.19780880906.
- Mori Y, Sato Y, Takamatsu S. Evolutionary analysis of the powdery mildew fungi using nucleotide sequences of the nuclear ribosomal DNA. Mycologia. 2000;92(1):74–93. doi: 10.1080/00275514.2000.12061132.
- Heluta V. Filogeneticheskie vzaimosvyazi mezhdu rodami erizifalnykh gribov i nekotorye voprosy sistematiki poryadka Erysiphales. Biologicheskii Zhurnal Armenii. 1988;41:351–358.
- Braun U, Shin H, Takamatsu S, et al. Phylogeny and taxonomy of Golovinomyces orontii revisited. Mycol Prog. 2019;18(3):335–357. doi: 10.1007/s11557-018-1453-y.
- Qiu P-L, Liu S-Y, Bradshaw M, et al. Multi-locus phylogeny and taxonomy of an unresolved, heterogeneous species complex within the genus Golovinomyces (Ascomycota, Erysiphales), including G. ambrosiae, G. circumfusus and G. spadiceus. BMC Microbiol. 2020;20(1):51. doi: 10.1186/s12866-020-01731-9.
- Bradshaw MJ, Braun U, Pfister DH. Phylogeny and taxonomy of the genera of Erysiphaceae, part 1: Golovinomyces. Mycologia. 2022;114(6):964–993. doi: 10.1080/00275514.2022.2115419.
- Braun U, Cook R. Taxonomic manual of the Erysiphales (powdery mildews). (CBS biodiversity series; no. 11). CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands; 2012. p. 1–707.
- Takamatsu S, Matsuda S, Niinomi S, et al. Molecular phylogeny supports a northern hemisphere origin of Golovinomyces (Ascomycota: Erysiphales). Mycol Res. 2006;110(9):1093–1101. doi: 10.1016/j.mycres.2006.07.005.
- Bradshaw MJ, Guan G-X, Nokes L, et al. Secondary DNA barcodes (CAM, GAPDH, GS, and RPB2) to characterize species complexes and strengthen the powdery mildew phylogeny. Front Ecol Evol. 2022;10:918908. doi: 10.3389/fevo.2022.918908.
- Gan P, Tsushima A, Hiroyama R, et al. Colletotrichum shisoi sp. nov., an anthracnose pathogen of Perilla frutescens in Japan: molecular phylogenetic, morphological and genomic evidence. Sci Rep. 2019;9(1):13349. doi: 10.1038/s41598-019-50076-5.
- Bakhshi M, Arzanlou M, Babai-Ahari A, et al. Novel primers improve species delimitation in Cercospora. IMA Fungus. 2018;9(2):299–332. doi: 10.5598/imafungus.2018.09.02.06.
- Berbee M, Pirseyedi M, Hubbard S. Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia. 1999;91(6):964–977. doi: 10.1080/00275514.1999.12061106.
- Bradshaw M, Tobin PC. Sequencing herbarium specimens of a common detrimental plant disease (powdery mildew). Phytopathology. 2020;110(7):1248–1254. doi: 10.1094/PHYTO-04-20-0139-PER.
- White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, et al., editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. p. 315–322.
- Artimo P, Jonnalagedda M, Arnold K, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40(W1):W597–W603. doi: 10.1093/nar/gks400.
- Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38(7):3022–3027. doi: 10.1093/molbev/msab120.
