Abstract
Pancratium maritimum L. (Amaryllidaceae) is a geophyte occurring in the Mediterranean region, from the Black Sea to part of the Atlantic coast. This plant is receiving much attention from the international scientific community due to its value as a bioindicator, the potential industrial value of its chemical compounds, and its use as a commercial ornamental plant. Plant morphometry and sequences of three plastid DNA regions (rbcL, matK, trnH-psbA) were used to assess the phenotypic and genetic variability of this taxon and its closest congeneric species (in particular Pancratium linosae, from the volcanic island of Linosa) in the Central Mediterranean (Sicily, Tunisia and surrounding islands). Pancratium maritimum and P. linosae cannot be distinguished based on morphological and genetic data and should belong to the same taxon. Our results also highlight a diversified gene pool in P. maritimum that is worth preserving. The lectotypes of the names Halmira stellaris, Pancratium angustifolium and Pancratium foetidum are here designated.
Introduction
Pancratium maritimum L. is one of the best known and most typical species of the Mediterranean littoral. In recent years, the level of attention given to this species has increased,because of its value as a bioindicator, the potential industrial value of its chemical compounds, and its use as a commercial ornamental plant (Abbassy et al. Citation1998; Berkov, Evstatieva, and Popov Citation2004; Bogdanova et al. Citation2009; El-Hadidy et al. Citation2011; Georgiev et al. Citation2011; Sanaa et al. Citation2013; Youssef and Frahm Citation1998). Alginic acid and derivatives were successfully extracted from the bulbs of this species (Sanaa et al. Citation2013). Several alkaloids with pharmacological activity were also found in P. maritimum. The most interesting of these was galanthamine, an acetylcholinesterase inhibitor (Berkov, Evstatieva, and Popov Citation2004). This species has also been thoroughly investigated from the biological (Eisikowitch and Galil Citation1971; Medrano, Guitián, and Guitián Citation2000), chemical (Ronsted et al. Citation2012), micro-morphological (Perrone et al. Citation2015), and genetic (Sanaa and Fadhel Citation2010; Sanaa et al. Citation2014; De Castro et al. Citation2012, Citation2014; De Felice et al. Citation2013; Di Maio and De Castro Citation2013) points of view.
Recently, many typical Mediterranean ecosystems have been severely altered by the increasing pressure of human activities and the consequences of global changes (de Montmollin and Strahm Citation2005; Sciandrello, Guglielmo, and Spampinato Citation2014). Among threatened habitats, sandy shores are one of the most endangered. Their severe decline is mainly the result of the human activities, tourism and construction development, which have modified the vegetation composition and the shorelines by alteration of the flow of water and sand deposition (Guarino et al. Citation2008). Moreover, the proliferation of invasive exotic species, such as Carpobrotus acinaciformis (L.) L. Bolus, has reduced the habitat available to native, less competitive species (Celesti-Grapow et al. Citation2010).
In the central Mediterranean, four species of Pancratium occur: Pancratium maritimum L., Pancratium linosae Soldano & F. Conti, Pancratium foetidum Pomel, and Pancratium illyricum L. (Conti et al. Citation2005; Euro+Med Citation2006). Pancratium maritimum and P. linosae are coastal plants, whereas P. foetidum and P. illyricum are rupicolous-heliophilous plants that also grow in the hinterland, up to 1350 m above sea level (Maire and Quézel Citation1959; Pignatti Citation1982). Pancratium maritimum, in contrast to most of the other species of Pancratium, has a wide distribution extending all over the Mediterranean, up to the Black Sea and to part of the Atlantic coast (Euro+Med Citation2006). In contrast, P. linosae is known only from the Pelagian Islands (De Castro et al. Citation2012). We therefore aimed to investigate the taxonomic independence of P. linosae within the area of distribution of P. maritimum.
In the context of development of a molecular database of the threatened flora of Sicily and surrounding areas (Giovino et al. Citation2015), the potential of the plastid markers, rbcL, matK and trnH-psbA, was assessed for Pancratium.
Material and methods
Pancratium was studied in the central Mediterranean, i.e. Sicily and Tunisia along a latitudinal transect from 35°N to 38°N. Specimens were collected in the field and voucher specimens were deposited in the Herbarium Mediterraneum Panormitanum (PAL) (Table ). Field identification was based on morphological characters in the mature stage. Individual plants were cultivated in the Botanical Garden of Palermo to exclude environmental variability, for subsequent comparative morphological analyses. For each population, character traits were measured for 10 individuals (five from Zembra due to the small size of this population) with 10 replicate measurements from each individual. Measurements were taken using an electronic calliper. The quantitative characters considered were: #1 Leaf length (cm); #2 Leaf width (cm); #3 Flowering stem length (cm); #4 No. of flowers per stem; #5 External length of tepals (cm); #6 Paracorolla total length (cm); #7 Paracorolla teeth length (cm). The means of the measures used for statistics are presented in the Supplemental data (File 1). According to Boyd (Citation2002) and Peruzzi et al. (Citation2015) these characters were subjected to a Principal Component Analysis, with the individuals a priori assigned to the eight populations (Figure ). Each character was also subjected to univariate analysis (analysis of variance or Kruskal–Wallis test with corrections for multiple comparisons, Pearson correlation coefficients, Tukey’s honest significant difference test and Bonferroni, respectively), using PAST version 3.06 (Hammer, Harper, and Ryan Citation2001; Hammer Citation2015). Pearson correlation coefficients (r) among the seven characters measured are presented in the Supplemental data (File 2). A discriminant analysis for the eight a priori groups recognized was performed. The scatter plot of specimens along the first two canonical axes is shown (Figure ). The range of each continuous numerical character was represented using box-and-whisker plots (Figure ).
Table 1. Synoptic table of the populations studied reporting the field identification, location, habitat characteristics and voucher details.
Figure 1. Principal components analysis based on the seven considered morphological characters, with groups corresponding to the eight populations. PC1: Eigenvalue 55.81, % variance 81.50; PC2: Eigenvalue 8.17, % variance 11.93. Yellow: Chebba; magenta: Zembra; red: Lampedusa; light green: Linosa 1; dark green: Linosa 2; brown: Cannatello; dark blue: Marsala, light blue: Balestrate.
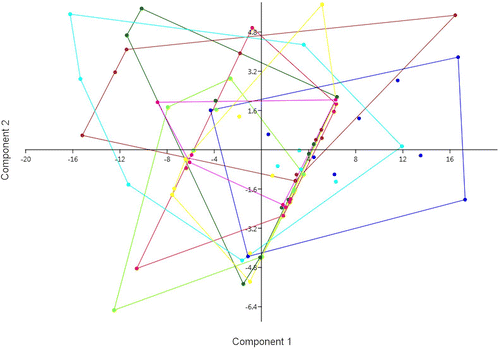
Figure 2. Discriminant analysis based on the seven considered morphological characters, with groups corresponding to the eight populations. Axis 1: Eigenvalue 0.40153, % variance 38.6; Axis 2 Eigenvalue 0.32117, % variance 30.88. Yellow: Chebba; magenta: Zembra; red: Lampedusa; light green: Linosa 1; dark green: Linosa 2; brown: Cannatello; dark blue: Marsala, light blue: Balestrate.
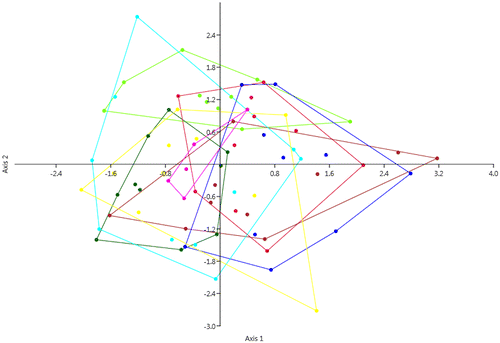
Figure 3. Plots of the six continuous numeric characters. For each sample, the 25–75% quartiles are drawn using a box. The median is shown with a horizontal line inside the box. The whiskers are drawn from the top of the box up to the largest data point less than 1.5 times the box height from the box, and similarly below the box. Outliers values are shown as stars. No significant differences among populations are highlighted. T(white): Overall; 1(yellow): Chebba; 2(magenta): Zembra; 3(red): Lampedusa; 4(light green): Linosa 1; 5(dark green): Linosa 2; 6(brown): Cannatello; 7(dark blue): Marsala, 8(light blue): Balestrate.
The barcoding approach was adopted in support of the morphological investigation. Multiple individuals for each taxon were used for molecular analysis. Plant material for DNA extraction consisted of young lyophilized leaves. Genomic DNA extraction was based on CTAB protocol for plant tissue (Doyle and Doyle Citation1987).
The three plastid barcoding regions rbcL, matK, trnH-psbA, were assessed by adopting polymerase chain reaction primers and conditions suggested by the Consortium for the Barcode of Life (CBOL) (Fazekas et al. Citation2012; Dunning and Savolainen Citation2010) (Table ). When making the choice of markers we considered the relevance of the compromise between the discrimination level supported by a marker and amplification and sequencing success (Chase et al. Citation2005, Hollingsworth et al. Citation2009). The choice of trnH-psbA, as an additional marker, appeared logical to discriminate morphologically close samples.
Table 2. Barcoding polymerase chain reaction primers adopted in this study.
Polymerase chain reaction amplifications were performed with the Gene®Amp PCR System 9700 (Applied Biosystems, Foster City, CA, USA). Products were purified and bi-directionally sequenced (Amersham Biosciences DYEnamic™ ET Terminator Cycle Sequencing Kits), according to the Sanger protocol for AB3730XL DNA Analyzer (Applied Biosystems). The resulting electropherograms were screened for errors and assembled into contigs using Sequencer software 4.10 (Gene Codes Corporation, Ann Arbor, MI, USA). The sequence alignments were carried out by MUSCLE and phylogenetic Neighbour-Joining. A tree was generated for molecular identification, based on a Kimura two-parameter model, using Mega 6 software (Kimura Citation1980; Saitou and Nei Citation1987; Tamura et al. Citation2013). The comparison included all new sequences generated, a subset of the most closely related sequences, and the significant BLAST results, downloaded from GenBank database (Table ). Sampling, morphological and molecular data generated in this investigation were submitted to the BOLD database under the dedicated project code FMED (www.boldsystems.org).
Table 3. Reference data of the newly generated sequences and the subset of the related sequences for each taxon considered in this study.
Results
The principal components analysis (Figure ) and the discriminant analysis (Figure ) indicated considerable overlap across the populations in this study. The cases correctly classified by discriminant analysis according to the groups assigned a priori were 36%. Univariate analysis of all the characters considered did not indicate any significant differences across populations. Morphological characters (Figure ) show continuous variation and do not fall into distinct population groupings.
All the barcoding fragments were successfully amplified and sequenced from the samples investigated. rbcL did not show any variation. Low rbcL polymorphism has been reported in numerous plant lineages (e.g. Chase et al. Citation2007; de Vere et al. Citation2012; Costion et al. Citation2011) and this marker was therefore discarded from subsequent analyses. Conversely, matK and trnH-psbA showed significant levels of genetic variation.
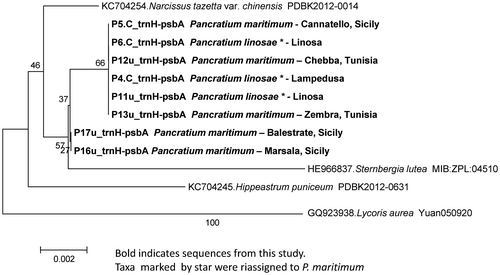
The phylogenetic tree, both of matK and trnH-psbA, showed concordant results, indicating no genetic divergence between the reference sequences of P. maritimum, recovered from GenBank, and those of the majority of the samples collected in this study (Figures and ). However, two Sicilian populations (taxa ID: P16u, P17u), originating from Balestrate and Marsala (north Sicily), were different on both markers.
Figure 4. Phylogenetic neighbour-joining tree generated from the most significant matK gene sequences listed in Table .
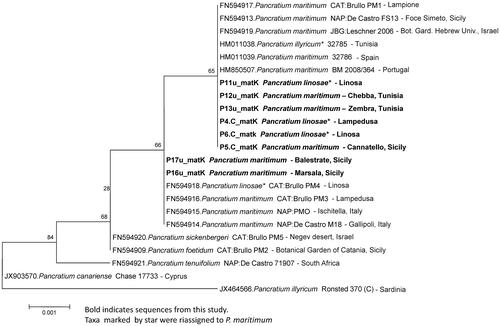
Figure 5. Phylogenetic neighbour-joining tree of the most significant trnH-psbA IGS sequences listed in Table .
No divergence was observed when comparing samples from plants in the in vivo collection.
Discussion
There was no statistically significant variation in morphological character traits of leaves, stems and flowers between the cultivated plants from the eight different locations, originally recognized as P. maritimum and P. linosae. This indicates that these plants belong to a single taxon. Considerations in Di Silvestro, Ardenghi, and Parolo (Citation2010), De Castro et al. (Citation2012) and Perrone et al. (Citation2015) on analysis of plants in wild populations, lead in the same direction, although the significant differences detected in the field provide evidence for high within-species phenotypic variability.
In spite of the overall low level of genetic variability observed among the sequences generated in this study, matK and trnH-psbA revealed a level of genetic variation that is high enough to support some considerations and help in clarifying some doubtful sequences associated with this group in GenBank. First, the matK sequence of P. maritimum from Marsala and Balestrate, matched precisely the reference sequence ascribed to P. linosae (ID: FN594918, voucher CAT: Brullo PM4). This result still argues for considering P. maritimum and P. linosae as phenotypic variants of the same taxon. Second, we also highlight the incorrect GenBank taxa assignment for the matK sequence referred to P. illyricum from Tunisia (Table , see ID: HM011038, voucher 32785). The phylogenetic tree showed this voucher specimen referable to P. maritimum (Figure ). Moreover P. illyricum is known to be endemic to Corsica and Sardinia, so the sampling location is rather unlikely to refer to this species.
The trnH-psbA intergenic spacer was used for the first time on Pancratium taxa. Quite surprisingly, it showed low molecular variation; even though it is known to be highly variable in complex groups (Chase et al. Citation2007; Chen et al. Citation2010; Hollingsworth, Graham, and Little Citation2011; De Mattia et al. Citation2011). Nevertheless, this fragment confirmed the variability of the two Sicilian samples (P16u, P17u), supported also by matK.
The evaluation of additional regions could represent a possibility for deeper investigations on the genetic variability of such taxa, in order to increase the resolution of molecular discrimination (Di Maio and De Castro Citation2013). Nonetheless, other molecular studies on the P. maritimum group, using further markers (i.e. trnL-trnF, trnL, ITS), also demonstrated a low level of genetic variation (Gage et al. Citation2011; De Castro et al. Citation2012). A population genetic study could help to assess gene flow between different locations, and identify differentiated gene pools (see below).
The following classification of the species of Pancratium occurring in the central Mediterranean is therefore proposed:
Pancratium maritimum L., Sp. Pl.: 291. 1753
= P. linosae Soldano & F. Conti, Annot. Checkl. Italian Vasc. Fl. 20. 2005. Type: homotypic with P. angustifolium Lojac.
= P. angustifolium Lojac., Fl. Sic. 3: 82. 1909., non M. Roem., Syn. Mon., 4 : 178. 1847. Typus: Lectotype here designated: Pancratium …. n. spec. Linosa si coltiva e si attenda che fiorisca [manu Tineo] / Pancratium angustifolium Lojac. [manu Lojacono] PAL 63913!
Typus: Lectotype: “Narcissus maritimus” in Morison, Pl. Hist. Univ., 2: 365, s. 4, t. 10, f. 28, 1680 designated by Wijnands in Jarvis & al. (ed.), Regnum Veg. 127 : 73 (1993).
Pancratium foetidum Pomel, Nouv. Mat. Fl. Atl. 253. 1874
= Pancratium collinum Coss. & Durieu, Ann. Sci. Nat., Bot., IV, 1: 228. 1854 [nom. nud.]
Typus: Lectotype here designated: Pancratium foetidum, Oran [manu Pome] / Pancratium foetidum Pomel, (type !), Oran, Pomel [manu Maire] MPU 005173!
Pancratium illyricum L., Sp. Pl.: 291. 1753
= Zouchia illyrica (L.) Raf. Fl. Tellur. 4: 22. 1838
= Almyra stellaris (Salisb.) Salisb., Trans. Hort. Soc. London 1: 336. 1812 [nom. illeg.]
= Halmira stellaris Parlatore, Nuov. Gen. Sp. Monocot.: 28. 1854 Typus: Lectotype here designated: Halmira stellaris Parl. Fl. It., Pancratium illyricum Linn., da Moris in Marzo1847, in apricis collinis: 8bri, 9bri Sardinia, FI fototeca3010B!
= Pancratium stellare Salisb., Trans. Linn. Soc. London 2: 74. 1794 [nom. illeg.]
Typus: Lectotype designated by Peruzzi et al. Citation2013: 828: Herb. Van Royen No. 897.326–441 (L).
Conservation
The morphological and genetic variability observed highlights the importance of preserving as many sites as possible in order to maintain a large gene pool. In fact, all genotypes resulting from local adaptation to a particular habitat are independent genetic units deserving conservation. Sandy shores of the Mediterranean are homogeneous habitats; they host species with broad distributions and are subject to continuous variations (Sànchez Doreste, Domina, and Caujapé-Castells Citation2005; Jimenez-Alfaro et al. Citation2015). In this case, as supposed by Picó and Van Groenendael (Citation2007) and by Geraci, Rimondo, and Troia (2009), it is reasonable to believe that fragmentation of the original population and the consequent geographical isolation of the sub-populations have promoted the genetic variation observed. The patchy genotypic distribution can be explained in terms of the seed dispersal capability of the plants and in terms of the importance of Mediterranean sea currents in dispersal processes, allowing the establishment of new colonies that are distant from the parent population (Balestri and Cinelli Citation2004; Westberg and Kadereit Citation2009; Raimondo et al. Citation2012; De Castro et al. Citation2014 Sanaa et al. Citation2015). Pancratium maritimum has a relatively very high dispersal capacity, giving rise to probable panmixy throughout the whole central Mediterranean as observed by Kadereit et al. (Citation2005) in other coastal species such as Cakile maritima Scop., Crithmum maritimum L. and Halimione portulacoides (L.) Aellen. The continuous decline of P. maritimum reduces the size of populations and their internal variability. It is therefore desirable to preserve P. maritimum populations in situ with a “dynamic” strategy that includes management of ecosystems, maintenance of gene flow, reintroduction of plants, environmental education, etc. Pancratium maritimum, taking into account its broad distribution, its distinctive appearance, and its iconic status, should be adopted as symbol plant of the sandy shore habitat in conservation initiatives.
Previous experiences of replant–transplant processes showed that overall performance of plants tends to be higher in populations in their native sites in comparison with populations replanted in non-native sites (Joshi et al. Citation2001; Bischoff et al. Citation2006). From this, it follows that it is important to know the distribution of genotypes in the field. This is necessary to be able to select the most suitable material for reintroduction in sites that have become too distant from the existing natural populations. This distance is related to the average dispersal distance of each species.
Ex situ conservation should be based on the collection of as large a number of seeds as possible from different populations in mainland and island habitats in order to conserve as much variability as possible. The use of this plant as an ornamental, from plants grown from seeds, and not bulbs dug from the wild, as generally happens, would increase the reserves of genetic material available without reducing the number of mature individuals in natural populations.
Conclusions
This work confirms the validity of the integrated approach linking molecular and morphological analysis, promoted by the CBOL, as the best solution for assigning a real meaning to the levels of nucleotide divergence observed and for developing an accurate universal database for molecular identification (Rougerie et al. Citation2009, Dunn Citation2003).
Pancratium maritimum is a highly variable species, from both the genotypic and phenotypic points of view, and this has allowed it to colonize sandy shores of different geologies and in some cases, rocky cliffs. This implies a concerted and intense effort to collect, conserve and reintroduce propagules from as many individuals and populations as possible to ensure the conservation of this species.
The distinction between P. maritimum and P. linosae is not supported by consistent morphological or molecular data. The samples of P. linosae from its locus classicus represent just one example of the broad intraspecific variation that can be found along the coast of the Mediterranean. The particular morphology observed on Linosa appears to be related to the nature of the extremely arid volcanic sands in which it grows. The same probably applies to the individuals of P. maritimum growing in rocky fissures on the islet of Lampione (CAT: Brullo PM1 in Figure and Table ), which show very similar morphological characters (Lo Cascio and Pasta Citation2012). In this case too, speciation by genetic drift does not need to be invoked, and character traits observed within this population are well within the range of the broad morphological and genetic variation observed in P. maritimum.
Notes on contributors
Antonio Giovino, PhD is a researcher at the Agricultural Research Council (CRA). The main focus of his research is on molecular studies and conservation of native higher plants. During the past years he studied the conservation biology of Mediterranean plants. Contribution: Molecular analysis and Morphometry.
Gianniantonio Domina, PhD is a researcher at the University of Palermo of Applied Botany, General Secretary of OPTIMA (the Organization for the Phyto-Taxonomic Investigation of the Mediterranean Area). The main focus of his research is about plant taxonomy and nomenclature. Contribution: Field work and Morphometry.
Giuseppe Bazan, PhD is Associate Professor at the University of Palermo of Applied Botany. The main focus of his research is about geographical information systems applied to botany and to plant conservation. Contribution: Morphometry
Patrizia Campisi, PhD is a researcher at the University of Palermo of Applied Botany. The main focus of her research is on conservation of higher plants and mosses, and on taxonomy of mosses. Contribution: Morphometry
Silvia Scibetta, PhD is an associated researcher at the Agricultural Research Council (CRA). The main focus of her research is on molecular analysis and taxonomy of higher plants and fungi. Contribution: Molecular analysis and Morphometry.
Supplemental data
Supplemental data for this article can be accessed at http://dx.doi.org/10.1080/12538078.2015.1089416.
Electronic supplementary file 1. Morphological characters (mean) used for the statistical analysis.
Electronic supplementary file 2. Pearson correlation coefficients (r) among the seven characters measured. All correlations were significant with p > 0.0001 (Student’s t-test) with the lone exception of External tepals length.
Supplementary_files.zip
Download Zip (13.6 KB)Acknowledgements
Our grateful thanks go to Dr Chira Nepi, curator of FI, for the digital loan of the exsiccata of Halmira stellaris and to Dr Sandro Lanfranco (University of Malta) for his careful critical reading of the manuscript.
Additional information
Funding
References
- Abbassy, M.A., and O.A. el-Gougary, S. el-Hamady, and M.A. Sholom. 1998. “Insecticidal, acaricidal and synergistic effects of soosan, Pancratium maritimum extracts and constituents.” Journal of the Egyptian Society of Parasitology 28: 197–205.
- Balestri, E., and F. Cinelli. 2004. “Germination and early-seedling establishment capacity of Pancratium maritimum L. (Amaryllidaceae) on coastal dunes in the North-Western Mediterranean.” Journal of Coastal Research 20: 61–770.
- Berkov, S., L. Evstatieva, and S. Popov. 2004. “Alkaloids in Bulgarian Pancratium maritimum L..” Zeitschrift für Naturforschung B 59c : 65–69.
- Bischoff, A., L. Cremieux, M. Smilauerova, C.S. Lawson, S.R. Mortimer, J. Dolezal, V. Lanta, et al. 2006. “Detecting local adaptation in widespread grassland species – the importance of scale and local plant community.” Journal of Ecology 94: 1130–1142.
- Bogdanova, Y., B. Pandova, S. Yanev, and M. Stanilova. 2009. “Biosynthesis of lycorine by in vitro cultures of Pancratium maritimum L. (Amaryllidaceae).” Biotechnology & Biotechnological Equipment 23: 919–922.
- Boyd, A. 2002. “Morphological Analysis of Sky Island populations of Macromeria viridiflora (Boraginaceae).” Systematic Botany 27: 116–126.
- Bruni, I., F. De Mattia, S. Martellos, A. Galimberti, P. Savadori, M. Casiraghi, P.L. Nimis and M. Labra. 2012. “DNA Barcoding as an Effective Tool in Improving a Digital Plant Identification System: A Case Study for the Area of Mt. Valerio, Trieste (NE Italy)”. PLoS ONE 7(9): e43256.
- Celesti-Grapow, L., A. Alessandrini, P.V. Arrigoni, S. Assini, E. Banfi, E. Barni, M. Bovio, et al. 2010. “Non-native flora of Italy: Species distribution and threats.” Plant Biosystems 144: 12–28.
- Chase, M.W., N. Salamin, M. Wilkinson, J.M. Dunwell, R.P. Kesanakurthi, N. Haidar, and V. Savolainen. 2005. “Land plants and DNA barcodes: short-term and long-term goals.” Philosophical Transactions of the Royal Society B: Biological Sciences 360: 1889–1895.
- Chase, M.W., R.S. Cowan, P.M. Hollingsworth, C. van den Berg, S. Madriñán, G. Petersen, O. Seberg, et al. 2007. “A proposal for a standard protocol to barcode all land plants.” Taxon 56: 295–299.
- Chen, S., D.K. Kim, M.W. Chase and J.H. Kim. 2013. “Networks in a large-scale phylogenetic analysis: reconstructing evolutionary history of Asparagales (Lilianae) based on four plastid genes.” PLoS ONE 8(3): e59472.
- Chen, S.L., H. Yao, J.P. Han, C. Liu, J.Y. Song, L. Shi, Y. Zhu, et al. 2010. “Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species.” PLoS ONE 5: e8613.
- Conti, F., G. Abbate, A. Alessandrini and C. Blasi. 2005. An annotated checklist of the Italian vascular flora. Roma: Palombi Editori.
- Costion, C., A. Ford, H. Cross, D. Crayn, M. Harrington, and A. Lowe. 2011. “Plant DNA barcodes can accurately estimate species richness in poorly known floras.” PLoS ONE 6: e26841.
- De Castro, O., S. Brullo, P. Colombo, S. Jury, P. De Luca, and A. Di Maio. 2012. “Phylogenetic and biogeographical inferences for Pancratium (Amaryllidaceae), with an emphasis on the Mediterranean species based on plastid sequence data.” Botanical Journal of the Linnean Society 170: 12–28.
- De Castro, O., A. Di Maio, G. Imparato, E. Véla, S. Barbarito, G. Sibilio, B. Menale et al. 2014. “Aggiornamenti e novità sulle conoscenze di Pancratium maritimum (Amaryllidaceae) [Updates and News on the knowledge on Pancratium maritimum (Amaryllidaceae)].” In: Floristica, Sistematica ed Evoluzione [Floristics, Systematics and Evolution], edited by L. Peruzzi, and G. Domina, pp. 39–40. Firenze: SBI.
- De Felice, B., F. Manfellotto, R. D’Alessandro, O. De Castro, A. Di Maio, and M. Trifuoggi. 2013. “Comparative transcriptional analysis reveals differential gene expression between Sand Daffodil tissues.” Genetica 141: 443–452.
- De Mattia, F., I. Bruni, A. Galimberti, F. Cattaneo, M. Casiraghi, and M. Labra. 2011. “A comparative study of different DNA barcoding markers for the identification of some members of Lamiaceae.” Food Research International 44: 693–702.
- de Vere, N., T.C.G. Rich, C.R. Ford, S.A. Trinder, C. Long, C.W. Moore, D. Satterthwaite, et al. 2012. “DNA barcoding the native flowering plants and conifers of Wales.” PLoS ONE 7: e37945.
- Di Maio, A., and O. De Castro. 2013. “Development and characterization of 21 microsatellite markers for Pancratium maritimum L. (Amaryllidaceae).” Conservation Genetics Resources 5: 911–914.
- Di Silvestro, D., N.G.M. Ardenghi, and G. Parolo. 2010. “Il Progetto FOR-MARE, FORMAzione e Ricerca scientifica in aree marine protette: stage di Ecologia marina e Geobotanica applicate” [The Project FOR-MARE, Formation and Scientific Research in marine protected areas: Stage of Applied Marine ecology and Geobotany]. In: XIX Congresso del Gruppo per l’Ecologia di Base “G. Gadio” [XIX Congress of the group for basic Ecology “G. Gadio”]. Olbia, 21–23 maggio 2010, Riassunti, 32.
- Doyle, J.J., and J.L. Doyle. 1987. “A rapid isolation procedure for small amounts of leaf tissue.” Phytochemical Bulletin 19: 11–15.
- Dunn, C.P. 2003. “Keeping taxonomy based in morphology.” Trends in Ecology & Evolution 18: 270–271.
- Dunning, L.T., and V. Savolainen. 2010. “Broad-scale amplification of matK for DNA barcoding plants, a technical note.” Botanical Journal of the Linnean Society 164: 1–9.
- Eisikowitch, D., and L. Galil. 1971. “Effect of wind on the pollination of Pancratium maritimum L. (Amaryllidaceae) by hawkmoths (Lepidoptera: Sphingidae).” Journal of Animal Ecology 40: 673–678.
- El-Hadidy, A., M. Abd El-Ghani, W. Amer, and R. Hassan. 2011. “Systematic Revision of the Genus Pancratium L. (Amaryllidaceae) in Egypt with a New Addition.” Notulae Scientia Biologicae 3: 24–38.
- Euro+Med. 2006. “Euro+Med PlantBase – the information resource for Euro-Mediterranean plant diversity.” Accessed October 9 2014. http://ww2.bgbm.org/EuroPlusMed/.
- Fazekas, A.J., M.L. Kuzmina, S.G. Newmaster, and P.M. Hollingsworth. 2012. “DNA Barcoding Methods for Land Plants.” Methods in Molecular Biology 858: 223–252.
- Gage, E., P. Wilkin, M.W. Chase, and J. Hawkins. 2011. “Phylogenetic systematics of Sternbergia (Amaryllidaceae) based on plastid and ITS sequence data.” Botanical Journal of the Linnean Society 166: 149–162.
- Georgiev, V., I. Ivanov, S. Berkov, and A. Pavlov. 2011. “Alkaloids biosynthesis by Pancratium maritimum L. shoots in liquid culture.” Acta Physiologiae Plantarum 33: 927–933.
- Geraci, A., F.M. Rimondo, and A. Troia. 2012. “Genetic diversity and local population structure in Ambrosina bassii (Araceae, Ambrosineae), a Mediterranean relict species.” Biochemical Systematics and Ecology 37: 737–746.
- Giovino, A., P. Marino, G. Domina, A. Scialabba, R. Schicchi, G. Diliberto, C. Rizza, and S. Scibetta. 2015. “Evaluation of the DNA barcoding approach to develop a reference dataset for the threatened flora of Sicily.” Plant Biosystems. doi:10.1080/11263504.2014.989285.
- Guarino, R., A. Guglielmo, F. Ronsisvalle, and S. Sciandrello. 2008. “Il progetto ECONET-COHAST: strategie per la conservazione degli habitat costieri.” Fitosociologia 44: 333–337.
- Hammer, Ø. 2015. “PAST 3.06.” Accessed May 25 2015. http://folk.uio.no/ohammer/past.
- Hammer, Ø., D.A.T. Harper, and P.D., Ryan. 2001. “PAST: Paleontological Statistics software package for education and data analysis.” Paleontologia Electronica 4: 1–9.
- Hollingsworth, P.M., and CBOL Plant Working Group. 2009. “A DNA barcode for land plants.” Proceedings of the National Academy of Sciences of the United States of America 106: 12794–12797.
- Hollingsworth, P.M., S.W. Graham, and D.P. Little. 2011. “Choosing and using a plant DNA barcode.” PLoS ONE 6: e19254.
- Jimenez-Alfaro, B., C. Marcenò, R. Guarino, and M. Chytry. 2015. “Regional metacommunities in two coastal systems: spatial structure and drivers of plant assemblages.” Journal of Biogeography 42: 452–462.
- Joshi, J., B. Schmid, M.C. Caldeira, P.G. Dimitrikopoulos, J. Good, R. Harris, A. Hector, et al. 2001. “Local adaptation enhances performance of common plant species.” Ecology Letters 4: 536–544.
- Kadereit, J.W., R. Arafeh, G. Somogyi, and E. Westberg. 2005. “Terrestrial growth and marine dispersal? Comparative phylogeography of five coastal plant species at a European scale.” Taxon 54: 861–876.
- Kimura, M. 1980. “A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences.” Journal of Molecular Evolution 16: 111–120.
- Lo Cascio, P., and S. Pasta. 2012. “Lampione, a paradigmatic case of Mediterranean island biodiversity.” Biodiversity Journal 3: 311–330.
- Maire, R., and P. Quézel. 1959. Flore de l’Afrique du Nord, 6. Paris: Paul Lechevalier.
- Medrano, M., P. Guitián, and J. Guitián. 2000. “Patterns of fruit and seed set within inflorescences of Pancratium maritimum (Amaryllidaceae): nonuniform pollination, resource limitation, or architectural effects?” American Journal of Botany 87: 493–501.
- de Montmollin, B. and W. Strahm. 2005. The Top 50 Mediterranean Island Plants: Wild plants at the brink of extinction, and what is needed to save them. B. de Montmollin and W. Strahm (eds), Gland and Cambridge: IUCN/SSC Mediterranean Islands Plant Specialist Group IUCN.
- Perrone, R., C. Salmeri, S. Brullo, P. Colombo, and O. De Castro. 2015. “What do leaf anatomy and micro-morphology tell us about thepsammophilous Pancratium maritimum L. (Amaryllidaceae) in response to sand dune conditions?” Flora 213: 20–31.
- Peruzzi, L., A. Santangelo, and C. E. Jarvis. 2013. “Lectotypification of Linnaean names in the Italian endemic flora.” Taxon 62: 827–829.
- Peruzzi, L., G. Astuti, A. Carta, F. Roma-Marzio, D. Dolci, F. Caldararo, F. Bartolucci, and L. Bernardo. 2015. “Nomenclature, morphometry, karyology and SEM cypselae analysis of Carduus brutius (Asteraceae) and its relatives.” Phytotaxa 202: 237–249.
- Picó, F.X., and J. Van Groenendael. 2007. “Large-scale plant conservation in European semi-natural grasslands: a population genetic perspective.” Diversity and Distributions 13: 920–926.
- Pignatti, S. 1982. Flora d’Italia, 1–3. Bologna: Edagricole.
- Raimondo, F.M., A. Scialabba, G. Zecca, F. Grassi, G. Casazza, and L. Minuto. 2012. “Genetic diversity in the endangered Sicilian endemic Brassica rupestris: Proposals for a conservation strategy.” Plant Biosystems 146: 847–856.
- Ronsted, N., M.R. Symonds, T. Birkholm, and S. Brogger Christensen. 2012. “Can phylogeny predict chemical diversity and potential medicinal activity of plants? A case study of Amaryllidaceae.” BMC Evolutionary Biology 12: 182.
- Rougerie, R., T. Decaëns, L. Deharveng, D. Porco, S.W. James, C.-H. Chang, B. Richard, et al. 2009. “DNA barcodes for soil animal taxonomy.” Pesquisa Agropecuária Brasileira 44: 789–801.
- Saitou, N., and M. Nei. 1987. “The neighbor-joining method: A new method for reconstructing phylogenetic trees.” Molecular Biology and Evolution 4: 406–425.
- Sanaa, A., and N.B. Fadhel. 2010. “Genetic diversity in mainland and island populations of the endangered Pancratium maritimum L. (Amaryllidaceae) in Tunisia.” Scientia Horticulturae 125: 740–747.
- Sanaa, A., A. Boulila, M. Boussaid, and N.B. Fadhel. 2013. “Alginic acid and derivatives, new polymers from the endangered Pancratium maritimum L.” Industrial Crops and Products 44: 290–293.
- Sanaa, A., A. Boulila, M. Boussaid, and N.B. Fadhel. 2014. “Pancratium maritimum L. in Tunisia: Genetic and chemical studies among the threatened populations.” Industrial Crops and Products 60: 75–78.
- Sanaa, A., S.B. Adib, A. Boulila, C. Messaoud, M. Boussaid, and N.B. Fadhel. 2015. “Modeling hydrochory effects on the Tunisian island populations of Pancratium maritimum L. using colored Petri nets.” Biosystems 129: 19–24.
- Sànchez Doreste, J. L., G. Domina, and J. Caujapé-Castells. 2005. “Genetic differentiation of three species of Matthiola (Brassicaceae) in the Sicilian insular system.” Plant Systematics and Evolution 253: 81–93.
- Schaefer, H., O.J. Hardy, L. Silva, T.G. Barraclough and V. Savolainen. 2011. "Testing Darwin's naturalization hypothesis in the Azores". Ecology Letters 14(4): 389–396.
- Sciandrello, S., G.A. Guglielmo, and G. Spampinato. 2014. “Spatial patterns and floristic composition of plant communities in coastal salt marshes of southeastern Sicily (Italy).” Acta Botanica Gallica 161: 99–109.
- Tamura, K., G. Stecher, D. Peterson, A. Filipski, and S. Kumar. 2013. “MEGA6: Molecular Evolutionary Genetics Analysis version 6.0.” Molecular Biology and Evolution 30: 2725–2729.
- Westberg, E., and J.W. Kadereit. 2009. “The influence of sea currents, past disruption of gene flow and species biology on the phylogeographical structure of coastal flowering plants.” Journal of Biogeography 36: 1398–1410.
- Youssef, D.T., and A.W. Frahm. 1998. “Alkaloids of the flowers of Pancratium maritimum.” Planta Medica 64: 669–670.
