Abstract
A pair of primers was designed to amplify the propylene alcohol dehydrogenase gene sequence based on the cDNA sequence of the tobacco allyl-alcohol dehydrogenase gene. All introns were sequenced using traditional polymerase chain reaction (PCR) methods and T-A cloning. The sequences from common tobacco (Nicotiana tabaccum L.) and rustica tobacco (Nicotiana rustica L.) were analysed between the third intron and the fourth intron of the propylene alcohol dehydrogenase gene. The results showed that the alcohol dehydrogenase gene is a low-copy nuclear gene. The intron sequences have a combination of single nucleotide polymorphisms and length polymorphisms between common tobacco and rustica tobacco, which are suitable to identify the different germplasms. Furthermore, there are some single nucleotide polymorphism sites in the target sequence within common tobacco that can be used to distinguish intraspecific varieties.
Introduction
Tobacco (Nicotiana sp.) is one of many plants currently used as models for molecular studies and is an economically important crop. The interspecific phylogenetic relationships within tobacco are complicated due to complex hybridization events within the tobacco lineage. DNA molecular markers have been used to identify germplasms and to accelerate the breeding cycle. The second generation of molecular markers that include random amplification of polymorphic DNA (RAPD), amplified fragment length polymorphism (AFLP) and simple sequence repeats (SSR) has been employed broadly to differentiate tobacco varieties,[Citation1–4] while a third generation of molecular markers, intron polymorphisms (IPs) based on single nucleotide polymorphisms (SNPs), has been used as well. Abundant noncoding sequences, introns, are interspersed in the eukaryote genome and have been shown to accumulate considerable nucleotide variations.[Citation5] At least half of the introns (>57,000) in tobacco are suitable for the exploitation of IP markers.[Citation6] In this work, the regional IPs of the tobacco propylene alcohol dehydrogenase gene were analysed interspecifically and intraspecifically to assist in the identification of the germplasms studied.
Materials and methods
The tobacco samples used were all provided by the Mudanjiang Tobacco Research Institute (). The genomic DNA was isolated from tobacco samples, using a plant genomic DNA extraction kit (TianGen Bio Inc., China). With a template from common tobacco named C1, a pair of primers was designed according to the mRNA for the tobacco propylene alcohol dehydrogenase gene (GenBank: AB036735.1). The two primers were designed to amplify the DNA for tobacco propylene alcohol dehydrogenase gene, and the sequences were as follows, C1-S: 5′-CAAAAGTTAAGAAAATGGC-3′, C1-A: 5′-AGTCCAGTGGCTTAACTC-3′. The PCR materials included 5 μL of 10 × PCR buffer, 4 μL of dNTP (2.5 mmol/L each), 1 μL of each primer (10 μmol/L), 1 μL of DNA template and 1.5 U of ex-Taq DNA polymerase (Takara Bio Inc., Tokyo, Japan). PCR was performed at 94 °C for 5 min, then 94 °C for 30 s, 58 °C for 30 s, 72 °C for 1 min for 30 cycles, with a hold at 72 °C for 20 min. The PCR products were sequenced after their detection on agarose/EB gels. The intron distribution was analysed according to the GT-AG rule in the target gene with reference to its cDNA sequence.
Table 1. Tobacco samples used in this study.
The specific primers were designed to target the boundary between the third intron and the fourth intron of the tobacco propylene alcohol dehydrogenase gene, and the IP sequences were identified as follows, IP-S: GTGCTGGAAGCAAAGAAAAG, IP-A: CAATTCCATCAGGGAAGTAC. The PCR materials included 5 μL of 10 × PCR buffer, 4 μL of dNTP (2.5 mmol/L each), 1 μL of each primer (10 μmol/L), 1 μL of the DNA template and 1.5 U of rTaq DNA polymerase (Takara Bio Inc., Tokyo, Japan).
PCR was performed at 94 °C for 5 min, then 94 °C for 30 s, 58 °C for 30 s, 72 °C for 1 min for 30 cycles, with a hold at 72 °C for 20 min. The PCR products were ligated to a pGEM-T vector (Promega Corporation, Madison, WI, USA) and sequenced on an ABI 3730 sequencer after electrophoretic detection in a 1.0% agarose/EB gel.
Results and discussion
The propylene alcohol dehydrogenase gene of sample C1 contained four introns and five exons that were 225, 178, 167, 83 and 379 bp long (Online Supplemental Appendix 1). The boundary sequence between the intron and exon segments were the sites of interest for which a primer was designed to target.
The target PCR product from the rustica tobacco (Nicotiana rustica) exhibited two bands when both primers, IP-S and IP-A, were used, while those of common tobacco (sun-shined tobacco and flue-cured tobacco) exhibited only one band on the 1.0% agarose/EB gel (). The results showed that these primers have the ability to differentiate common tobacco from rustica tobacco, and that there are two members of the propylene alcohol dehydrogenase gene with length polymorphisms in rustica tobacco.
Figure 1. Agarose/EB gel electrophoresis results of the amplification of the target region in sun-shined tobacco samples (a); flue-cured tobacco samples (b); and rustica tobacco samples (c).
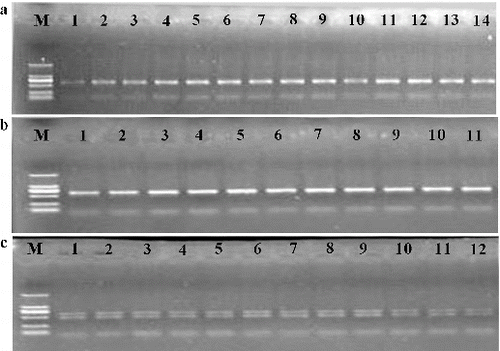
The sequence results also displayed two copies of propylene alcohol dehydrogenase gene with length polymorphisms in common tobacco in a 3.0% agarose/EB gel, as two bands of the PCR product were able to be differentiated from each other (data not shown).
In the common tobacco, the shorter fragment was 559-bp long in the target region, and the two introns found were 223 and 253 bp. There were no length differences in this fragment among the 12 samples sequenced, and the consensus sequence was as follows:
GTAAGTTCCATTGCGTTGGATCTTCATAGTGTTGAATCTATCTTCATTCTTCGTTATGTTAATCATTGTGCTCCGTCCACCATCAAACCCAAAGGTTGGGGTTGGGGTTGGGGTTAGGGTTGGGTGGGTTATGCCTTTGGGGGGAAAGAGAGGTTTTAGTTTAATAGACCATGATGTTTAAAACTCTTGTTATTTAATGTCTCTTTTTTATTCATTTCCAAAGGTTGATCTGTTGAAGAGCAAATTTGGTTTTGACGAGGCTTTTAACTATAAAGAGGAGCAGGATTTAAATGCAGCTTTGAAGAGGTGAGTAATCTTACTTCCTGCTACAAAACAATCTACCTATTGTACAAAAGCAGTTAAAGATGTTCTGTGAAAAGTATGATGATATTATGTAGATAGTGTGAAACTTATAGAAAGACTTTAAGATATGCCTACGAAAACCGAGGAACATGGATCATTTCAAATCTGGCTTTTCATGAATATGGAACCCCATTCACTAGATTTTAGTGCGAAATTCCTAGACTTGAAGTGACTGGAAATACTGGTTTGTTATTAG
In the common tobacco, the longer sequence obtained was between 588 and 591-bp long in the target region, and the two introns were either between 194 and 197 bp or 311 bp in length. The sequences from 13 samples were analysed with the software ClusterW2 (www.ebi.ac.uk/Tools/clustalw2/). The results indicated that there were two transitions and one transversion in addition to three gaps within the third intron. The two transitions and one transversion were also present within the fourth exon, but no amino acid replacement had occurred. There were four transitions and one transversion within the fourth intron as well (data shown in Online Supplemental Appendix 2). These polymorphic sites within the third and fourth intron may function as references to distinguish the varieties of common tobacco studied here.
In the rustica tobacco, the shorter sequences were 442-bp long in the target region, and the two introns found were 195 and 164 bp in length. No length polymorphisms were found in this fragment among the 12 samples sequenced, and the consensus sequence obtained was as follows:
GTAAGTTCCTTCATAGTGTTGAAGCTCTCTTCATTCTTCGTTATGTTAATTATTGTGCTCCGTCCTCCATCAAACCCAAAGGTTGGGGTTTTGTGGGTTATGAGGAGGAGAAAGGGGTATACCTTTGGGGGGGAAGAGATGTTTTAGTTTAATAGACCATGATGTTTAACTCTCTTTTTATATTCATTTCCAAAGGTTGATCTGTTGAGGAACAAATTTGGTTTTGACGAAGCTTTTAACTATAAAGAGGAGCAGGACTTAAATGCAGCTTTGAAGAGGTGAGTAATCTTACTACTTGGCCTGGTGTTAGAGAAGCAGTTAGCAACAGCTTTTCATCAATATTTCCATAGAATGATATTTATAATGGAAGTCCCAAAGTAAAGATGTTCAGTGAAAAGTATGCCTGTGAAAACTGAGGGAAATAATACTGGTTTCTTATTAG
In the rustica tobacco, the longer sequence was 583-bp long in the target region, and the two introns found were 220 and 280 bp in length. No polymorphisms were found in this fragment among the 12 samples sequenced as well, and the consensus sequence was as follows:
GTAAGTTCCATTGCGTTGGATGTTCATAGTGTTGAAGCTATCTTCATTCTTCGTTATGTTAATCATTGTGTTCTGTCCACCGTCAAACCCAAAGGTTGGGGTTTTGTGGGTTATGAGGAGGAGAAAGGTATACCTTTGGGGGGAAGAGATGTTTTAGTTTAATAGACCATGATGTTTAACTATCTTTTTATTTAATGTCTCTTTTTATTCATTTCCAAAGGTTGATCTGTTGAAGAGCAAATTTGGTTTTGACGAAGCTTTTAACTATAAAGAGGAGCAGGACTTAAATTCAGCTTTGAAGAGGTGAGTAATCTTACTTCCTGTTAGAAAACAATCTACCTATTGTAACAAACTTGCCCTGGTGTTAGAGAAGCAGTTAGCAACAGCTTTTCCTTTTCATGTAAACGGAATCATCGATATTTCCATAGAATGATATTTATTCCTTGAACTGAAAAATAGAAGTCCCAAAGTAAAGATGTACAGTGAAAAGTATGAAGATTGTGTGAAAATTATAGAAAAGACTTTAGGATATGCGTATGAAAACTGAGGGAAATTCCTCCCTGGAATACTGGTTTCTTATTAG
In this study, the tobacco propylene alcohol dehydrogenase gene proved to be a low-copy nuclear gene. It could have formed as a result of divergent evolution after gene duplication, or by a parallel evolutionary process between the two sets of genomes, as the tobacco plants tested were all allotetraploid. Although the PCR product for the allyl-alcohol dehydrogenase gene of sample C1 was sequenced successfully, this does not mean that only one copy of the target gene is present in common tobacco. There were obvious intron length polymorphisms between common tobacco and rustica tobacco, which were mostly due to different modes of evolution of the two kinds of tobacco.
It is thought that the maternal ancestor of common tobacco was Nicotiana sylvestris.[Citation7–9] It is also thought that its paternal ancestor was derived from either N. tomentosiformis,[Citation10–15] N. otophora or from introgressive hybridization of N. otophora and N. tomentosiformis within the subgenus of common tobacco.[Citation16,17] It has been shown that N. rustica originated from a natural chromosome duplication event after ancestral hybridization of N. paniculata and N. undulate.[Citation18,19] Yang et al. [Citation20] divided 48 tobacco samples into two categories, the N. rustica L. group and Nicotiana tabaccum L. group, after an analysis that used AFLP methods coupled with unweighted pair group method with arithmetic mean analysis, which was consistent with our results in this work. Furthermore, the two categories of tobacco can be distinguished using only one pair of primers according to the results of this work.
Conclusions
The two copies of the propylene alcohol dehydrogenase gene found in tobacco each contained four introns and five exons. The length polymorphisms of the intron existed intraspecifically as well as interspecifically. The primers produced here, IP-S and IP-A, were able to easily differentiate common tobacco from rustica tobacco in a 1% agarose gel. Some SNPs of the intron within common tobacco were found, and may be useful in the identification of varieties of this species.
Supplemental data
Supplemental data and research materials for this article can be accessed at http://dx.doi.org/10.1080/13102818.2014.907651.
tbeq_a_907651_sm4733.doc
Download MS Word (43.5 KB)Additional information
Funding
References
- Chang AX, Qu YS, Jia XH. Optimalization of tobacco RAPD reaction system and studies on polymorphic marker of tobacco varieties. Chin Tob Sci. 2004;2:9–13. Chinese.
- Rossi L, Zhao BD. A method of distinguishing different tobacco varieties amplified fragment length polymorphism (AFLP). Acta Tabacaria Sinica. 1999;4:5. Chinese.
- Xu MH, Zheng MH, Liu GT. Genetic diversity in cultivars of Nicotiana tabacum revealed by RAPD markers. J Agri Biotechnol. 1998;6:281–285. Chinese.
- Yang BC, Xiao BG, Chen XJ. Genetic diversity of flue-cured tobacco varieties based on ISSR markers. Hereditas. 2005;27:753–758. Chinese.
- Wang XS, Zhao XQ, Wu WR. Genome-wide investigation of intron length polymorphisms and their potential as molecular markers in rice (Oryza sativa L.). DNA Res. 2005;12:417–427.
- Zhang XW, Wang ZD, Liu YH. IP marker and its application strategy in molecular breeding of tobacco. J Anhui Agri Sci. 2008;36:3147–3148, 3159. Chinese.
- Aoki S, Ito M. Molecular phylogeny of Nicotiana (Solanaceae) based on the nucleotide sequence of the matK gene. Plant Biol. 2000;2:316–324.
- Bland MM, Matzinger DF, Levings CS. Comparison of the mitochondrial genome of Nicotiana tabacum with its progenitor species. Theor Appl Genet. 1985;69:535–541.
- Yukawa M, Tsudzuki T, Sugiura M. The chloroplast genome of Nicotiana sylvestris and Nicotiana tomentosiformis: complete sequencing confirms that the Nicotiana sylvestris progenitor is the maternal genome donor of Nicotiana tabacum. Mol Genet Genomics. 2006;275:367–373.
- Komarnitskiĭ SI, Komarnistkiĭ IK, Cox A, Parokonnyĭ AS. Molecular phylogeny of the nuclear 5.8S ribosomal RNA genes in 37 species of Nicotiana genus. Genetika. 1998;34:883–889.
- Lim KY, Matyášek R, Lichtenstein CP, Leitch AR. Molecular cytogenetic analyses and phylogenetic studies in the Nicotiana section Tomentosae. Chromosoma. 2000;109:245–258.
- Murad L, Lim KY, Christopodulou V, Matyasek R, Lichtenstein CP, Kovarik A, Leitch AR. The origin of tobacco's T genome is traced to a particular lineage within Nicotiana tomentosiformis (Solanacese). Am J Bot. 2002;89:921–928.
- Petit M, Lim KY, Julio E, Poncet C, de Borne FD, Kovarik A, Leitch AR, Grandbastien MA, Mhiri C. Differential impact of retrotransposon populations on the genome of allotetraploid tobacco(Nicotiana tabacum). Mol Genet Genomics. 2007;278:1–15.
- Riechers DE, Timko MP. Structure and expression of the gene family encoding putrescine N-methyltransferase in Nicotiana tabacum: new clues to the evolutionary origin of cultivated tobacco. Plant Mol Biol. 1999;41:387–401.
- Sperisen C, Ryals J, Meins F. Comparison of cloned genes provides evidence for intergenomic exchange of DNA in the evolution of a tobacco glucan endo-1,3-β-glucosidase gene family. P Natl Acad Sci U.S.A. 1991;88:1820–1824.
- Chase MW, Knapp S, Cox AV, Clarkson JJ, Butsko Y, Joseph J, Savolainen V, Parokonny AS. Molecular systematics, GISH and the origin of hybrid taxa in Nicotiana (Solanaceae). Ann Bot-London. 2003;92:107–127.
- Kenton A, Parokonny AS, Gleba YY, Bennett MD. Characterization of the Nicotiana tabacum L. genome by molecular cytogenetics. Mol Gen Genet. 1993;240:159–169.
- Dadejová M, Lim KY, Soucková-Skalická K, Matyásek R, Grandbastien MA, Leitch A, Kovarík A. Transcription activity of rRNA genes correlates with a tendency towards intergenomic homogenization in Nicotiana allotetraploids. New Phytol. 2007;174:658–668.
- Lim KY, Matyasek R, Kovarik A, Fulnecek J, Leitch AR. Molecular cytogenetics and tandem repeat sequence evolution in the allopolyploid Nicotiana rustica compared with diploid progenitors N. paniculata and N. undulata. Cytogenet Cell Genet. 2005;109:298–309.
- Yang YC, Zhou QM, Yin HQ, Zhu LS. Studies on genetic diversity and relationship in tobacco germplasms by AFLP analysis. Scientia Agricultura Sinica. 2006;39:2194–2199. Chinese.
