Abstract
The mammalian major histocompatibility complex (MHC) plays important roles in pathogen recognition and disease resistance. In the present study, the coding sequence and the 5′- and 3′-untranslated regions of MHC class II DR alpha chain (the DRA gene) from rare gayal and gaytle were cloned and analyzed to dissect structural and functional variations. The nucleotide and amino acid sequences for the DRA genes in gayal (Bofr-DRA) and gaytle (Bofr × BoLA-DRA) were almost identical to those for cattle and yak (99%). Compared to yak, two amino acids substitutions in the signal peptide (SP) domain for gayal were found within all Bos animals. Except for only one replacement in the amino acid within the α2 domain of the DRA protein in gayal, the additional residues were highly conserved across the species investigated. The 20 peptide-binding sites (PBS) of Bofr-DRA and Bofr × BoLA-DRA were essentially reserved in the α1 domain among all species investigated. The lesser degree of substitution in Bofr-DRA is concordant with the concept that the DRA gene is highly conserved among all mammals. The very high degree of conservativity of the DRA gene among ruminants, including gayal, suggests its recent evolutionary separation.
Introduction
The immune system is a function of biological structures and processes within an organism that protects against diseases by recognizing and responding to pathogens and antigens.[Citation1] The mammalian major histocompatibility complex (MHC) is a group of closely linked and highly variable genes and gene clusters.[Citation2] The cattle MHC, which is called bovine leukocyte antigen (BoLA), plays a key role in the defense against infectious diseases and regulates antigen presentation by encoding proteins that take part in the innate and adaptive immune responses to intruding pathogens.[Citation3,4] The MHC genes are associated with susceptibility or resistance to many diseases for cattle and sheep.[Citation5,6] Thus, the MHC genes could be indicated as primary candidates for the investigation of genetic variation for the purpose of marker-assisted breeding.
The gayal or mithun (Bos frontalis) is a rare semi-wild bovine species naturally inhabiting Indo-China.[Citation7,8] It has a chromosome complement of 2n = 58,[Citation9–11] which differs from those of their counterparts such as cattle (B. taurus, 2n = 60) and gaur (B. gaurus, 2n = 56).[Citation12] Gayal often consumes tree and bamboo leaves, grasses, reeds and other local plants and shows a very wide range of adaptations under the harsh conditions which range from cold to tropical regions.[Citation13,14] Moreover, the quality of meat, including tender and muscle fibre diameter, from gayal is better than in local cattle.[Citation15,16] Therefore, the hybrid between gayal (♂) and cattle (♀), which is also called gaytle (gayal × cattle), presents better meat traits than cattle and can meet the diversity demand for consumers. Due to the remoteness of their habitats, as well as socio-political and ecological factors, gayal is one of the least studied ruminants.[Citation17–20]
Until now, the MHC-DRA gene has been cloned and sequenced in European cattle [Citation21] and zebu,[Citation22] yak and yakow,[Citation23,24] buffaloes,[Citation25] goats,[Citation26] sheep,[Citation22,Citation27] donkeys,[Citation28] pigs,[Citation29] mice,[Citation30] cats [Citation31] and macaques.[Citation32] So far, the MHC-DRA from gayal and gaytle has not been deposited in the GenBank database (http://www.ncbi.nlm.nih.gov/). These native animals are more common sources of meat and milk than cattle in Indo-China. They are considered to be less susceptible than cattle to some endemic diseases.[Citation33,34] In the present study, we have isolated and characterized full-length cDNAs from gayal and gaytle and compared those with the sequences of MHC-DRA from different animal species in order to inspect the genetic factors for disease susceptibility and resistance.
Materials and methods
Samples collection, RNA extraction and first-strand cDNA synthesis
Liver RNA samples from three mature gayals (B. frontalis) and three mature gaytles (B. frontalis × B. taurus) were collected at the slaughter house, Gongshan County (Yunnan Province, China). The samples were snap frozen and kept in liquid nitrogen during storage and transportation, and then stored at −80 °C. The total RNA was isolated using a Total RNA Extraction Kit (Beijing Tiangen Biotech Co., Ltd., China). DNase I treatment of the total RNA was performed before the cDNA was constructed using RevertAid™ First Strand cDNA Synthesis Kits (Fermentas Inc., USA) following the manufacturer's instructions.
Primers and PCR amplification of the DRA gene
The MHC-DRA genes from gayal and gaytle were isolated using forward primer (5′-CGAGACACCGAAGAAGAAAAT-3′) and reverse primer (5′-GGAGGGAAAACCAATACAAGAA-3′), which were also successfully used to amplify DRA sequences from buffalo, yak and yakow.[Citation20,21] Polymerase chain reaction (PCR) was conducted using Bioer Life Express Thermocycler (Bioer Technology Co., Ltd., China) using 2 μL of cDNA template, 12.5 μL of 2× PCR Power Mix (Beijing Zoman Biotechnology Co., Ltd., China), 2 μL of 10 pmol/μL of each primer and 8.5 μL of double distilled water in a total reaction volume of 25 μL. The PCR conditions started with an initial denaturation temperature of 94 °C for 4 min, followed by 36 cycles of the following steps: 94 °C for 1 min, 56 °C for 1 min and 72 °C for 1 min, with a final extension of 10 min at 72 °C. Visualization of amplified products was performed on agarose gel stained with ethidium bromide. The PCR products were sequenced bi-directionally using an ABI373X DNA analyzer (Applied Biosystems Inc.) at the Sango Biotechnology Company (Shanghai, China).
Sequences analysis
The cDNA sequence prediction was conducted using the GenScan software (http://genes.mit.edu/GENSCAN.html).[Citation35] The theoretical isoelectric point (pI) and molecular weight (Mw) of the putative DRA proteins were computed using the Compute pI/Mw Tool (http://www.expasy.org/tools/pi_tool.html).[Citation36] The DNAStar v5.2.2 program and MEGA v5.0 software package with the neighbour-joining method [Citation37] were used to conduct sequences alignment and to construct a phylogenetic tree.
Results and discussion
Identification and characterization of the Bofr-DRA and Bofr × BoLA-DRA
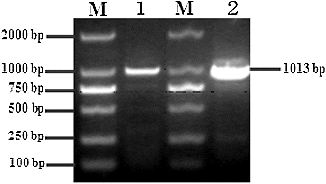
The resulting PCR products of 1013 bp for both Bofr-DRA (gayal: B. frontalis) and Bofr × BoLA-DRA (gaytle: B. frontalis × B. taurus) genes were obtained through reverse transcription PCR (RT-PCR) using liver cDNA as a template (). The fragment of 972 bp including the complete coding region (762 bp) and parts of the 5′- and 3′-untranslated regions (27 and 183 bp, respectively) of the MHC-DRA gene was effectively sequenced and deposited into the GenBank database with accession numbers KF981723 and KF981724, respectively. Compared with the bovine MHC-DRA (GenBank accession number D37956), the complete coding region of Bofr-DRA and Bofr × BoLA-DRA genes (762 bp) encoded a polypeptide of 253 amino acids with a predicted Mw of 28,425 and 28,407 Da (pI 5.41), respectively.
Sequence alignment and comparison
presents the nucleotide sequences and amino acid alignments. The Bofr-DRA and Bofr × BoLA-DRA genes had the closest similarity (99%) to the DRA gene of cattle, zebu, yakow and yak, with high level of similarity with the respective gene in buffalo, goat, sheep, pig, dog, macaque, human, cat and mouse. In addition to the signal peptide (SP), other functional domains, including the α1, α2, connecting peptide (CP), transmembrane (TM) and cytoplasmatic (CY) regions, are highly conserved, particularly the sites associated with biological function in these DRA genes.[Citation25] Importantly, the SP domain has been reported to have the most variable region of the molecule across the species.[Citation25] However, there was high similarity among Bofr-DRA, Bofr × BoLA-DRA genes and those of other ruminants.
Table 1. Sequence similarity comparison at the nucleotides and amino acids (within parentheses) sections for the signal peptide (SP), α1, α2 and connecting peptide/transmembrane/cytoplasmatic (CP/TM/CY) domains of the DRA genes from different species compared to those for gayal (B. frontalis, up) and gaytle (B. frontalis × B. taurus, down).
The entire SP, α1, α2 and CP/TM/CY domains from DRA molecules from gayal and gaytle were almost identical to those of cattle, except the mutation in position 138 in the amino acid chain within the α2 domain (P. M138L, ). However, compared with yak, there was minimal mutation of the Bofr-DRA and Bofr × BoLA-DRA genes, with only four single nucleotide substitutions within the coding regions, resulting in two amino acid replacements in the sixth and seventh position within the SP domain (p. V6S and p. P7Q).
Figure 2. Alignment of the protein encoded by the DRA genes in gayal (Bos frontalis), gaytle (B. frontalis × B. taurus), European cattle (B. taurus, D37956), zebu cattle (Bos indicus, FM986338), yak (Bos grunniens, JQ911700), Chinese yakow (B. grunniens × B. taurus, JQ347519), buffaloes (Bubalus bubalis, DQ016629), goats (Capra hircus, AB008754), sheep (Ovis aries, M73983), macaques (Macaca mulatta, EF208826), humans (Homo sapiens, NM_019111), pigs (Sus scrofa, M93028) and mice (Mus musculus, NM_010381). A hyphen (-) indicates amino acid identity and an asterisk (*) indicates a gap inserted for maximum alignment. Arrows (↓) within the green column indicate the amino acid positions constituting part of peptide-binding site (PBS). A circle (О) with a blue column indicates a conserved site for T-cell receptor interaction and putative N-linked glycosylation sites are underlined. A rectangle sign (![]()
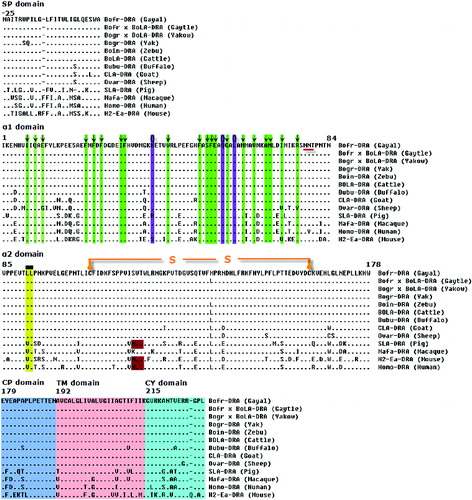
The 20 amino acids which are located in positions 7, 9, 11, 22, 24, 26, 31, 32, 43, 51, 53, 54, 55, 58, 62, 65, 68, 69, 72 and 76 of the peptide-binding site (PBS), as reported by Soen et al.,[Citation38] were highly conserved within the α1 domain (). Compared to other MHC class II gene families such as the DRB, DQA and DQB, which are highly variable at the PBS,[Citation39–42] the DRA gene appears to be highly conserved for residues forming the PBS region in different mammals, including primates. It seems that selection pressure has led to conservation for these specific functional regions in the DRA gene. However, there was evidence of variability in the first domain exon (α1) of DRA of ruminants.[Citation22,Citation43] These results could emphasize the complexity and function of the DRA gene, particularly in the PBS region. The cysteine residues that build an interchain disulphide bond between positions 107 and 163 were also stable and conserved. In gayal, gaytle and other animals including cattle, zebu, yak, yakow, buffaloes, sheep, goats and macaques, the DRA molecules have only one potential N-linked glycosylation site (78–80 amino acids) in the α1 domain as opposed to two that occur in pigs, humans and mice, with the second site located in the α2 domain (118–120 amino acids marked by the reddish box in ).[Citation21,Citation26] The deprivation of one glycosylation site in the α2 domain for gayal and other ruminants such as cattle and yak is due to the replacement of an asparagine residue with serine. It is not known whether the glycosylation site in the α2 domain may have an important role in the immune response and the response against specific antigens, although its lack presents a specific molecular characteristic of ruminant DRA genes. There were no substitutions in other important functional sites such as T lymphocytes receptor recognition site (positions 39, 57 and 60) and CD4 molecule-binding site (positions 91–92) in gayal, gaytle, yak, Chinese yakow, zebu, cattle, buffaloes, sheep and goats.
Phylogenetic analysis based on neighbour-joining method
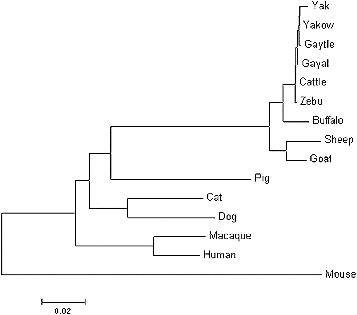
From the phylogenetic tree analysis (), it appears that the DRA genes of ruminant and non-ruminant species evolved independently, since the species they originate from have split into different branches over the evolutionary time. It is also apparent that yak and cattle are most closely related species to gayal.
Figure 3. Phylogenetic tree for the DRA genes from gayal, gaytle, European cattle, zebu, yak, Chinese yakow, buffaloes, goats, sheep, macaques, humans, pigs, cats, dogs and mice.
In the present study, the loss of mutation in Bofr-DRA and Bofr × BoLA-DRA is concordant with the DRA gene being conserved within ruminants and all mammalian species. As a result, this could be used for the identification of shared antigenic sites of similar pathogens or the recognition of very specific antigens which may be common to most of the species. Moreover, the close similarity of the DRA gene among ruminants, including gayal, may be ascribed to recent separation in evolutionary process and/or similar selection pressure which the ruminants have suffered during evolution.
Conclusions
The DRA genes of gayal and gaytle have been isolated and characterized for the first time, thereby enlarging cognizance of the MHC-DRA in rare ruminants. It points out that the Bofr-DRA gene is highly conserved, particularly in the first domain exon as in most mammals. In the future, it would be more interesting to dissect Bofr-DRB, which probably contains most of the polymorphism within the MHC-DR molecule.
Additional information
Funding
References
- Lazzaro BP, Little TJ. Immunity in a variable world. Philos Trans R Soc Lond B Biol Sci. 2009;364(1513):15–26. doi:10.1098/rstb.2008.0141
- Trowsdale J. “Both man & bird & beast”: comparative organization of MHC genes. Immunogenetics. 1995;41(1):1–17. doi:10.1007/BF00188427
- Lewin H, Russel G, Glass E. Comparative organization and function of the major histocompatibility complex of domesticated cattle. Immunol Rev. 1999;167:145–158. doi:10.1111/2Fj.1600-065X.1999.tb01388.x
- Luís C, Cothran EG, Oom MM, Bailey E. Major histocompatibility complex locus DRA polymorphism in the endangered Sorraia horse and related breeds. J Anim Breed Genet. 2005;122(1):69–72.
- Dukkipati VSR, Blair HT, Garrick DJ, Murray A. ‘Ovar-Mhc’ – ovine major histocompatibility complex: role in genetic resistance to diseases. NZ Vet J. 2006;54(4):153–160. doi:10.1080/00480169.2006.36689
- Morris K. The feline major histocompatibility complex. Univ Sydney Undergrad Res J. 2009;1(1):75–97.
- Mondal M, Dhali A, Rajkhowa C, Prakash BK. Secretion patterns of growth hormone in growing captive mithuns (Bos frontalis). Zoolog Sci. 2004;21(11):1125–1129. doi:10.2108/zsj.21.1125
- Rajkhova S, Arma DKS, Rajkhova C. Seroprevalence of Toxoplasma gondii antibodies in captive mithuns (Bos frontalis) from India. Vet Parasitol. 2006;135(3–4):369–374.
- Chi J, Fu B, Nie W, Wang J, Graphodatsky AS, Yang F. New insights into the karyotypic relationships of Chinese muntjac (Muntiacus reevesi), forest musk deer (Moschus berezovskii) and gayal (Bos frontalis). Cytogenet Genome Res. 2005;108(4):310–316. doi:10.1159/000081520
- Qu KX, He ZX, Nie WH, Zhang JC, Jin XD, Yang GR, Yuan XP, Huang BZ, Zhang YP, Zan LS. Karyotype analysis of mithun (Bos frontalis) and mithun bull × Brahman cow hybrids. Genet Mol Res. 2012;11:131–140. doi:10.4238/2012.January.19.1
- Xi DM, Wu M, Fan YY, Liu Q, Leng J, Gou X, Mao HM, Deng WD. Polymorphisms of the insulin-like growth factor-binding protein 3 gene (IGFBP3) in gayal (Bos frontalis). Gene. 2012;497(1):98–102. doi:10.1016/j.gene.2012.01.051
- Kamalludin MH. Cytogenetic studies of Malayan Gaur (Bos gaurus hubbacki), Sahiwal-Friesian cattle and their hybrid backcrosses. [Master's thesis]. Malaysia: Senate of Universiti Putra Malaysia; 2009 [Abstract].
- Xi DM, Wanapat M, Deng WD, He TB, Yang ZF, Mao HM. Comparison of gayal (Bos frontalis) and Yunnan Yellow Cattle (Bos taurus): in vitro dry matter digestibility and gas production for a range of forages. Asian-Aust J Anim Sci. 2007;20(8):1208–1214.
- Zhao KD, Ou CH, Huang YL, He TB. [Rare animal germplasm resources in Yunnan Province: present situation and counter measures of preservation and research on Dulong cattle (Bos frontalis)]. J Yellow Cattle Sci. 2003;29(2):71–74. Chinese. doi:10.3969/j.issn.1001–9111.2003.02.021
- Ge CR, Tian YB, Chen T, Wu Y. Studies on the meat feature of gayal (Bos frontalis). Sci Agric Sin. 1996;29(2):75–78. Chinese.
- Giasuddin M, Huque KS, Alam J. Reproductive potentials of gayal (Bos frontalis) under semi-intensive management. Asian-Aust J Anim Sci. 2003;16(3):331–334.
- Mohan M, Chandan R, Prakash BS. Development and validation of a highly sensitive economic enzyme immunoassay for prolactin determination in blood plasma of mithun (Bos frontalis) and its application during milk let down and cyclicity. Anim Reprod Sci. 2007;99(1–2):182–195. doi:10.1016/j.anireprosci.2006.05.003
- Md-Zain BM, Majid S, Rosli MKA, Yaakop S. Phylogenetic position of selembu and other cattle via paternal marker comparison. Life Sci J. 2014;11(12):42–46.
- Gao RC, Huang ZX, Wu CF, Xu B, Wang XY. Culture-independent analysis of microflora in Gayals (Bos frontalis) feces. Afr J Biotechnol. 2010;19(9):2774–2788.
- Sanyal S, Das PK, Ghosh PR, Das K, Vupru KV, Rajkhowa C, Mondal M. Electrocardiogram of clinically healthy mithun (Bos frontalis): variation among strains. Vet Med Int. 2010:790310. doi:10.4061/2010/790310
- Aida Y, Kohda C, Morooka A, Nakai Y, Ogimoto K, Urao T, Asahina M. Cloning of cDNA and the molecular evolution of bovine MHC class II DRA gene. Biochem Biophys Res Commun. 1994;204(1):195–202. doi:10.1006/bbrc.1994.2444
- Ballingall KT, Rocchi MS, McKeever DJ, Wright F. Trans-species polymorphism and selection in the MHC class II DRA genes of domestic sheep. PLoS One. 2010;5(6):e11402. doi:10.1371/journal.pone.0011402
- An TW, Zhang H, He JW, Feng HY, Luo YZ, Han JL. Polymorphic but highly conserved Bogr-DRA gene in yak (Bos grunniens). Anim Genet. 2012;43(2):237–238. doi:10.1111/j.1365–2052.2011.02256.x
- Sun Y, Zheng H, Xi D, Zhang X, Du M, Pu L, Lin M, Yang Y. Molecular characteristics of the MHC-DRA genes from yak (Bos grunniens) and Chinese yakow (Bos grunniens × Bos taurus). Int J Immunogenet. 2014;41(1):69–73. doi:10.1111/iji.12072
- Sakaram D, Niranjan SK, Kumar S, Naskar S, Deb SM, Mitra A, Sharma A, Sharma D. cDNA characterization and molecular analysis of buffalo MHC class II gene, DRA (Bubu-DRA). J Appl Anim Res. 2010;37(1):73–76. doi:10.1080/09712119.2010.9707097
- Takada T, Kikkawa Y, Yonekawa H, Amano T. Analysis of goat MHC class II DRA and DRB genes: identification of the expressed gene and new DRB alleles. Immunogenetics. 1998;48(6):408–412. doi:10.1007/s002510050452
- Fabb SA, Maddox JF, Gogolin-Ewens KJ, Baker L, Wu MJ, Brandon MR. Isolation, characterization and evolution of ovine major histocompatibility complex class II DRA and DQA genes. Anim Genet. 1993;24(4):249–255. doi:10.1111/j.1365–2052.1993.tb00307.x
- Vranova M, Alloggio I, Qablan M, Vyskocil M, Baumeisterova A, Sloboda M, Putnova L, Vrtkova I, Modry D, Horin P. Genetic diversity of the class II major histocompatibility DRA locus in European, Asiatic and African domestic donkeys. Infect Genet Evol. 2011;11(5):1136–1141. doi:10.1016/j.meegid.2011.04.010
- Hirsch F, Germana S, Gustafsson K, Pratt K, Sachs DH, Leguern C. Structure and expression of class II alpha genes in miniature swine. J Immunol. 1992;149(3):841–846.
- Subramanian S, Yim YS, Liu K, Tus K, Zhou XJ, Wakeland EK. Epistatic suppression of systemic lupus erythematosus: fine mapping of Sles1 to less than 1 Mb. J Immunol. 2005;175(2):1062–1072.
- Yuhki N, O’Brien SJ. Nature and origin of polymorphism in feline MHC class II DRA and DRB genes. J Immunol. 1997;158(6):2822–2833.
- O’Conner SL, Blasky AJ, Pendley CJ, Becker EA, Wiseman RW, Karl JA, Hughes AL, O’Connor DH. Comprehensive characterization of MHC class II haplotypes in Mauritian cynomolgus macaques. Immunogenetics. 2007;59(6):449–462. doi:10.1007/s00251–007-0209-7
- Rajkhowa S, Rajkhowa C, Rahman H, Bujarbaruah KM. Seroprevalence of infectious bovine rhinotracheitis in mithun (Bos frontalis) in India. Rev Sci Tech Off Int Epizoot. 2004;23(3):821–829.
- Renukaradhya GJ, Rajasekhar M, Raghavan R. Prevalence of infectious bovine rhinotracheitis in southern India. Rev Sci Tech Off Int Epizoot. 1996;15(3):1021–1028.
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268(1):78–94.
- Bjellqvist B, Hughes GJ, Pasquali C, Paquet N, Ravier F, Sanchez JC, Frutiger S, Hochstrasser D. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis. 1993;14(10):1023–1031. doi:10.1002/elps.11501401163
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425.
- Soen Y, Chen DS, Kraft DL, Davis MM, Brown PO. Detection and characterization of cellular immune responses using peptide-MHC microarrays. PLoS Biol. 2003;1(3):e65. doi:10.1371/journal.pbio.0000065
- Kamath PL, Getz WM. Adaptive molecular evolution of the major histocompatibility complex genes, DRA and DQA, in the genus Equus. BMC Evol Biol. 2011;11:128. doi:10.1186/1471-2148-11-128
- Niranjan SK, Deb SM, Sharma A, Mitra A, Kumar S. Isolation of two cDNAs encoding MHC-DQA1 and -DQA2 from the water buffalo, Bubalus bubalis. Vet Immunol Immunopathol. 2009;130(3–4):268–271. doi:10.1016/j.vetimm.2009.02.006
- Sena L, Schneider MP, Brenig BB, Honeycutt RL, Honeycutt DA, Womack JE, Skow LC. Polymorphism and gene organization of water buffalo MHC-DQB genes show homology to the BoLA DQB region. Anim Genet. 2011;42(4):378–385. doi:10.1111/j.1365-2052.2010.02157.x
- Sun YK, Xi DM, Li GZ, Hao TT, Chen YH, Yang YA. Genetic characterization of MHC class II DQB exon 2 variants in gayal (Bos frontalis). Biotechnol Biotech Equip. 2014;28(5):827–833. doi:10.1080/13102818.2014.960787
- Sena L, Schneider MP, Brenig B, Honeycutt RL, Womack JE, Skow LC. Polymorphisms in MHC-DRA and -DRB alleles of water buffalo (Bubalus bubalis) reveal different features from cattle DR alleles. Anim Genet. 2003;34(1):1–10. doi:10.1046/j.1365-2052.2003.00920.x