Abstract
Fusarium graminearum and F. culmorum are phytopathogenic species causing scab and root rot diseases in all small grain cereals worldwide including Turkey. In this study, resistance levels to geneticin (G418) of 14 F. graminearum and 24 F. culmorum isolates collected from cereals were determined. Fungal cultures were grown on potato dextrose agar medium supplemented with 0, 25, 50, 75 and 100 µg/mL of G418. Minimum inhibitory concentration was determined as 25 µg/mL. As a result, it was concluded that all isolates were highly sensitive to G418. Plasmid pFA6-kanmx4 containing geneticin resistance gene (kanmx) was introduced singly or co-electroporated with pEGFP75 plasmid, containing GFP gene, into fungal protoplast cultures obtained with lytic enzyme. Transformants were grown in media including 25 µg/mL G418. Transformation frequencies were 2.8 and 1.8 transformant per µg plasmid for F. graminearum and F. culmorum isolates, respectively. Transformation process was also confirmed by spectrofluorimetric assay. Relative fluorescence unit values in co-transformants were calculated as 1.87 ± 0.04 for F. graminearum and 2.26 ± 0.08 for F. culmorum. The results obtained from the study gave information about antibiotic resistance levels of two Fusarium species in Turkey. Moreover, it was shown that pFA6-kanmx4 plasmid was a suitable vector, which can be used in genetic manipulation studies of these two fungal species in particular suppression of endogenous and/or the expression of exogenous genes.
Introduction
Several strategies have been used in agriculture, in order to defend against phytopathogenic Fusarium species.[Citation1,Citation2] Development of resistant crop varieties to pathogen is one of the most common strategies. However, the process takes a long time. Also, developed varieties could have unfavourable agronomic traits. Applying the fungicides against pathogens is another approach to be used. Unfortunately, pathogen specific fungicides are absent. Therefore, there are some efforts for developing Fusarium isolates sensitive to fungicides.[Citation2,Citation3] The most natural approach is to provide the antagonistic effects of microorganisms on control of Fusarium.[Citation4,Citation5] Also, genetic manipulation of pathogen has been carried out in order to fight it.[Citation6,Citation7] The antibiotic resistance levels of a particular fungus will be used in genetic manipulation studies and should be tested according to designed transformation vectors.
Limited number of antibiotics, including amoxicillin, cefazolin, chloramphenicol, moxifloxacin, tobramycin and geneticin and hygromycin B, has been tested in studies associated with genetic manipulation of Fusarium genus.[Citation7,Citation8] Among them, hygromycin B resistance is well characterized in Fusarium. But no information has been available with respect to geneticin: G418 antibiotic resistance. Geneticin is an aminoglycoside antibiotic which inhibits the elongation step of translation in both prokaryotic and eukaryotic organisms. Therefore, it is not suitable for a cure. So it has been used as a selection marker in molecular studies.[Citation9,Citation10]
Vector-based gene manipulation approaches including gene disruption/deletion, gene silencing and/or over-expression have been effectively used for transformation of fungi by exogenous genes.[Citation6,Citation11,Citation12] In fungal transformation studies, a polyethylene glycol (PEG)-mediated gene transfer method applied to fungal protoplast cultures has been well examined and efficient strategies have been developed in some species of Ascomycota.[Citation6,Citation7,Citation13–15] However, different strategies gave different results on their stability and efficiency of fungal transformation. Electroporation-mediated gene transfer to fungal protoplasts is an alternative strategy.[Citation16] Plasmid vectors used for transformation should include at least marker genes, reporter gene and proper promoter and/or terminator regions and replication origins. Since selection of transformants is generally carried out by using antibiotics, plasmid vectors should carry antibiotic resistant genes. Hygromycin B is the most preferred antibiotic in the selection of transformants in Ascomycota. Therefore, most of plasmid vectors contain hygromycin B resistance gene (hygromycin B phosphotransferase; hph). Fungus susceptible to antibiotic might have an ability to eventually develop resistance to antibiotics overtime as well as in nature.[Citation8] Hence, effectiveness of different antibiotics and their related marker genes should be examined in gene manipulation studies. Geneticin:G418 resistance gene has been isolated from Micromonospora rhodorangea.Citation[Citation17] The gene can be used in fungal transformation studies by ligation of different plasmid vector scaffold. G418 included in plasmid vectors such as pFA6-kanmx4 could be used.
Determination of resistance levels of 38 isolates belonging to F. graminearum and F. culmorum to geneticin:G418 antibiotic was the major aim of this study. Transformation of two isolates belonging to two species by plasmid vectors containing G418 resistance gene via electroporation was also purposed.
Materials and methods
Fungal isolates, media and G418 resistance
Totally 14 F. graminearum and 24 F. culmorum single spore isolates obtained from scabby barley, maize and wheat were used in the determination of antibiotic resistance whereas transformation was carried out with only one F. graminearum (4F) and one F. culmorum (20F) isolate in this study (). Fungal isolates were grown on potato dextrose agar (PDA) plates for 10 days at 25 °C. Liquid cultures were also grown on potato dextrose broth (PDB) for 10 days at 120 rpm and 25 °C. The level of Geneticin:G418 resistance of isolates were evaluated by addition of G418 with various concentrations at 0, 25, 50, 75 and 100 µg/mL to PDA and PDB medium. Growth was determined via measurement of dry weight of mycelia and observing condiospore production.
Table 1. Fungal isolates used in this study.
Protoplast preparation
Mycelia harvested from fungal cultures grown on PDB medium were used in protoplast production. Novozyme 234 (Novo Industries, Denmark), Driselase (Sigma, Germany) and lytic enzyme (Sigma, Germany) obtained from Trichoderma sp. were used in the elimination of the cell walls. Enzymes were dissolved in digestion buffer (10 mmol/L Na2HPO4, 1.2 Mg2SO4, pH 5.8). Driselase, novozyme and lytic enzyme were added to liquid cultures with final concentration of 20 mg/mL. Six hours after enzyme addition, cultures were filtrated through Whatman paper grade 1 (Sigma, Germany). Two-fold volume of STC buffer (20% sucrose, 25 mmol/L Tris-HCl, pH 7.5, 25mM CaCl2) was added to the filtrate and samples were centrifuged at 800 × g for 10 minutes. Protoplasts were precipitated and then suspended by addition of STC buffer and they were used immediately in fungal transformation or stored for longer times at –70 °C.[Citation14,Citation18] The number of protoplasts were calculated and then diluted at final concentrations of 1 × 105 for transformation studies.
Plasmid vectors
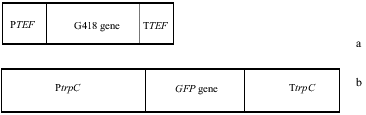
pFA6-kanmx4 and pEGFP75 plasmid vectors were used in this study. While pFA6-kanmx4 was provided from Dr Bedia Palabiyik, Department of Molecular Biology and Genetics, Faculty of Science, Istanbul University, pEGFP75 was a kind gift of Dr Hitoshi Nakayashiki from Plant Pathology and Microbiology Laboratory, Graduate School of Agricultural Sciences, Kobe University ( A, B).
Figure 1. Diagrams of plasmid vector used in this study. (a) 1.3 kb fragment of pFA6-kanmx4 plasmid and (b) 2.6 kb fragment of pEGFP75 plasmids. PTEF: A. gossypii TEF promoter, TTEF: A. gossypii TEF terminater, PtrpC: A. nidulans tryptophane promoter, TtrpC: A. nidulans tryptophane terminator regions.
Fungal transformation
Protoplasts at 1 × 105 cells/mL and plasmid vectors (pFA6-kanmx4 and pEGFP75) were used in electroporation mediated transformation. Each plasmid was transferred into protoplasts as singly or multi. 390 µL of protoplast culture and 10 µL of plasmid (1 µg/µL) were mixed in sterile microtubes and samples were chilled on ice for 20 minutes. Samples were transferred to electroporation cuvette and electric pulse comprising of 800 V and 0.8 µs was carried out. Samples were immediately transferred to ice and kept for 20 minutes. Protoplast cultures were transformed with pFA6-kanmx4 plasmid and were diluted in molten medium with 1.2 mol/L sorbitol. After centrifugation at 2000 x g for 10 minutes protoplasts were inoculated to PDA medium including 25 µg/mL of G418. Transformation frequency was calculated as a number of transformants per µg plasmid DNA.[Citation16,Citation18–20]
Selection of transformation
Confirmation of transformation was also carried out by co-introducing pFA6-kanmx4 with plasmid pEGFP75 [Citation19] containing GFP gene, tryptophane promotor (PtrpC) and terminator (TtrpC). Co-transformation was detected by fluorescence microscopy (Leica-MX100, Germany) and spectrofluorometic measurements obtaining relative fluorescence unit values (RFU) at 485/535 nm excitation/emission wawelengths.[Citation15,Citation21] Chromosomal DNA was extracted from co-transformants by using commercial kit based on the establish CTAB protocol (Macherey–Nagel, Germany). PCR primers GFPF/GFPR (5′- ATGGTGAGCAAGGGCGAGGA-3′/5′- TTACTTGTACAGCTCGTCCA-3′) and GF/GR (5′-TGGGAAAAGAGAAGACACACG-3'/5′-GGACTGAACTCCCCTAGACA) targeting GFP and G418 resistance genes, respectively, were used in PCR analysis. PCR conditions were maintained as described by Wang et al. [Citation14].
Results and discussion
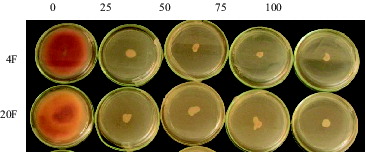
In order to determine G418 antibiotic resistance levels of Fusarium species, isolates were maintained on PDA and PDB medium for 10 days. All control groups were grown on medium without antibiotic (). However, none of fungal isolates were grown on medium supplemented with antibiotic at various concentrations as 25, 50, 75 and 100 µg/mL. Minimum inhibitory concentration (MIC) of G418 was thus set at 25 µg/mL in all F. graminearum and F. culmorum isolates.
Figure 2. 10-day-old cultures of (A) F. graminearum 4F and (B) F. culmorum (20F) isolates grown on PDA medium supplied with G418 at various concentrations as 0, 25, 50, 75 and 100 µg/ml.
Approximately, 5 mg fresh mycelia were used in protoplast preparation. Protoplast concentrations were calculated as 5 × 107 cells/mL with lytic enzyme applications (). However, 5 × 106 protoplasts were obtained from cultures supplemented with Novozyme 234 and Driselase. It was shown that lytic enzyme was more effective than Novozyme 234 and Driselase for preparation of protoplast cultures.
Figure 3. 20X capture of protoplasts obtained from mycelia by incubation with lytic enzyme for 6 hours. Bar indicates 50 µm length.

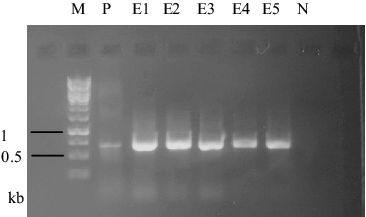
Fungal transformation was achieved by electroporation mediated-introduction of pFA6-kanmx4 plasmid including G418 resistance gene with Ashbya gossypii TEF promoter and terminator regions. Transformation was tested with protoplasts belonging to F. graminearum and F. culmorum. 1 × 105 protoplasts of these fungal isolates were transformed with pFA6-kanmx4 plasmid. Isolates transformed with G418 resistance gene were efficiently grown on PDA + 25 µg/mL G418 medium. Transformation frequency was 2.8 and 1.8 transformant per µg plasmid for F. graminearum and F. culmorum isolates, respectively. For the confirmation of the process, co-transformation of pFA6-kanmx4 with pEGFP75 plasmid was carried out to protoplast cultures. Mycelia of co-transformants were investigated under fluorescence microscopy and mycelia with GFP fluorescence were detected (). Moreover, RFU values in co-transformants were in the ranges of 1.87 ± 0.04–2.26 ± 0.08. GFP and G418 resistance gene fragments of 720 and 715 bp, respectively, were amplified from DNA molecules of transformants (). These data showed that the co-transformation method was more successful than single transformation method in the two Fusarium species. Similarly, Wiebe et al. [Citation18] and Scherm et al. [Citation7] reported high levels (5 colony/µg DNA) of co-transformation efficiency in Fusarium species. Possible reasons of high transformation efficiency obtained of this study can be as follows: (1) efficiency of co-transformation depends on species and transformation conditions. (2) Introduction of genes carried by plasmid to related region occurs independently from transformation. (3) Also, insertion of one gene into a genome could mediate that of other genes.[Citation22–24]
Figure 4. Fluorescence microscopy (20X capture) analysis of mycelia obtained from pEGFP75 transformed fungus.
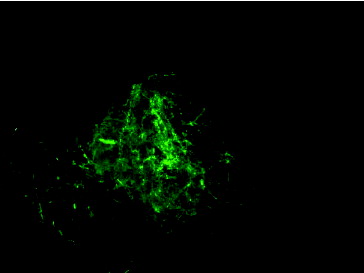
Figure 5. 720 bp DNA bands amplified from 5 pEGFP75 transformed colonies and positive control (pEGFP75 plasmid). M: 1 kb DNA ladder, N: negative control, P: positive control, E1-6: transformant samples.
Geneticin:G418 resistance profiles and electroporation mediated gene transfer efficiencies of Turkish Fusarium isolates were investigated in this study. Results obtained from this study showed that all fungal isolates were highly sensitive to G418. Sakaguchi et al. [Citation25] showed that Trichosporon cutaneum strains were at different hygromycin B-resistant levels. Similarly, Penicillium roqueforti and Ophiostoma piceae strains with different antibiotic resistance levels were reported by Durand et al. [Citation19] and Wang et al. [Citation14] Generally, hygromycin B has been selected as a marker gene in genetic transformation studies in Ascomycota. But, a great variation in the sensitivity to this antibiotic is present in fungi.[Citation8] Thus, according to the output of this study, G418 could be used as an effective selective marker in molecular genetic studies.
In genetic transformation studies associated with Ascomycota, protoplast/spheroplast cultures have commonly been used as compared to spore cultures. In addition, PEG-mediated transformation is widely used. Wiebe et al. [Citation18] reported that highest protoplast yield was obtained with Driselase application. In this study, protoplast cultures were obtained with three different agents and highest number of protoplast were obtained with lytic enzyme. Moreover, they obtained transformation efficiency in F. graminearum species at five transformants per µg DNA by PEG transformation. Higher levels of transformation efficiency were obtained by the electroporation-mediated gene transfer to protoplast cultures of Fusarium species in this study. Since transformation frequency in species belonging to Ascomycota showed high levels of variation ranging from 0.1 to 30 colony per μg DNA, different approaches for genetic transformation could be applied in fungal species. Co-transformation was also confirmed to be a stable and efficient transformation method in Fusarium species by electroporation. pAN7-1 containing hph gene or pEGFP75 plasmid with GFP gene are of approximately 6 kb sizes. The large sizes for plasmid could be problematic in fungal transformation.[Citation21,Citation26,Citation27] Moreover, resistance to hygromycin B is variable in fungal species. Thus, plasmid pFA6-kanmx4 of 3.6 kb size could be useful in fungal transformation studies as a primary selectable marker since it has relatively small length. In this study, it was shown that toxicity of geneticin antibiotic was high for Fusarium species. Therefore, high level of MIC values was obtained even at the low antibiotic concentrations. For that reason, it was revealed that G418 gene carried by pFA6-kanmx4 could be useful as a selectable marker for transformation studies.
Conclusion
This study reports on the geneticin resistance levels and the effectiveness of electroporation-mediated gene transfer of Fusarium isolates from Turkey. Results showed that fungal isolates were highly sensitive to this antibiotic. Additionally, plasmid vector including G418 gene was successfully transformed into protoplast cultures as singly and multi. The antibiotic resistance levels of Fusarium isolates which could be used in further gene silencing and/or over expression studies were tested. Findings of this study showed that geneticin antibiotic G418 could be utilized in practical applications of genetic transformation studies in Fusarium species.
Acknowledgements
Authors are grateful to Dr Berna Tunali, Department of Plant Protection, Agricultural Faculty, Samsun Ondokuz Mayis University for providing fungal material. We also thank Dr Bedia Palabiyik, Department of Molecular Biology and Genetics, Faculty of Science, Istanbul University and Dr Hitoshi Nakayashiki, Plant Pathology and Microbiology Laboratory, Graduate School of Agricultural Sciences, Kobe University for providing pFA6-kanmx4 and pEGFP75 plasmids.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernardo A, Bai G, Guo P, Xiao K, Guenzi AC, Ayoubi P. Fusarium graminearum -- induced changes in gene expression between Fusarium head blight-resistant and susceptible wheat cultivar. Functional Integr Genomic. 2007;7:69–77.
- Kimura M, Tokai T, Takahashi-Ando N, Ohsato S, Fujimura M. Molecular and genetic studies of Fusarium trichothecen pathways gene and evolution. Biosci Biotech Biochemistry. 2007;71:2105–2123.
- Bai G, Shaner G. Management and resistance in wheat and barley to Fusarium head blight. Annu Rev Phytopathol. 2004;42:135–161.
- Bello GMD, Monaco CI, Simon MR. Biological control of seedling blight of wheat caused by Fusarium graminearum with beneficial rhizosphere microorganisms. World J Microb Biotechnol. 2002;18(7):627–636.
- Altinok HH, Dikilitas M, Yildiz HN. Potential of Pseudomonas and Bacillus isolates as biocontrol agents Fusarium Wilt of Eggplant. Biotechnol Biotechnological Equipment. 2013;27(4):3952–3958.
- McDonald T, Brown D, Keller NP, Hammond TM. RNA silencing of mycotoxin production in Aspergillus and Fusarium species. Mol Plant Microbe Interactions. 2005;18(6):539–545.
- Scherm B, Orru M, Balmas V, Spanu F, Azara E, Delogu G, Hammond TM, Keller NP, Migheli Q. Altered trichothecene biosynthesis in TRI6-silenced transformants of Fusarium culmorum influences the severity of crown and foot rot on durum wheat seedlings. Mol Plant Pathol. 2011;12(8):759–771.
- Day S, Lalitha P, Haug S, Fothergill AW, Cevallos V, Vijayakumar R, Prajna NV, Acharya NR, McLeod SD, Lietman TM. Activity of antibiotics against Fusarium and Aspergillus. Br J Ophthalmology. 2009;93(1):116–119.
- Franke CA, Rice CM, Strauss JH, Hruby DE. Neomycin resistance as a dominant selectable marker for selection and isolation of vaccinia virus recombinants. Mol Cell Biol. 1985;5(8):1918–1924.
- Zhou H, Chen YQ, Du YP, Qu LH. The Schizosaccharomyces pombe mgU6-47 gene is required for 2΄-O-methylation of U6 snRNA at A41. Nucleic Acids Res. 2002;30(4):894–902.
- Brown DW, Dyer RB, McCormick SP, Kendra DF, Plattner RD. Functional demarcation of the Fusarium core trichothecene gene cluster. Fungal Genet Biol. 2004;41:454–462.
- Frandsen RJ, Nielsen NJ, Maolanon N, Sørensen JC, Olsson S, Nielsen J, Giese H. The biosynthetic pathway for aurofusarin in Fusarium graminearum reveals a close link between the naphthoquinones and naphthopyrones. Mol Microbiol. 2006;61:1069–1080.
- Smalley EB, Lin B. Research on fungal growth stimulants for domestic animals in China. Biotechnol Biotechnological Equipment. 1990;4–5(6):48–54.
- Wang HL, Kim SH, Siu H, Breuil C. Transformation of sapstaining fungi with hygromycin B resistance plasmids pAN7-1 and pCB1004. Mycological Res. 1999;103(1):77–80.
- Nakayashiki H, Hanada S, Quoc NB, Kadotani N, Tosa Y, Mayama S. RNA silencing as a tool for exploring gene function in ascomycete fungi. Fungal Genet Biol. 2005;42:275–283.
- Richey MG, Marek ET, Schardl CL, Smith DA. Transformation of filamentous fungi with plasmid DNA by electroporation. Phytopathol. 1989;79:844–847.
- Waitz JA, Sabatelli F, Menzel F, Moss JEL. Biological activity of antibiotic G-418, a new Micromonospora-produced aminoglycoside with activity against protozoa and helminthes. Antimicrob Agents Chemother. 1974;6(5):579–581.
- Wiebe MG, Novakova M, Miller L, Blakebrough ML, Robson GD, Punt PJ, Trinci APJ. Protoplast production and transformation of morphological mutants of the Quorn2 myco-protein fungus, Fusarium graminearum A3/5, using the hygromycin B resistance plasmid pAN7-1. Mycological Res. 1997;101(7):871–877.
- Durand N, Reymond P, Ftvre M. Transformation of Penicillium roqueforti to phleomycinand to hygromycin B-resistance. Curr Genet. 1991;19:149–153.
- Diez BR. Strategies for the transformation of filamentous fungi. J Appl Microbiol. 2002;92:189–195.
- Kadotani N, Nakayashiki H, Tosa Y, Mayama S. RNA silencing in the phytopathogenic fungus Magnaporthe oryzae. MPMI. 2003;16 (9):769–776.
- Depicker A, Herman L, Jacobs A, Schell J, Montagu VM. Frequencies of simultaneous transformation with different T-DNAs and their relevance to the Agrobacterium/plant cell interaction. Mol Genet Genomics. 1985;201:477–484.
- Wernars K, Goosen T, Wennekes BM, Swart K, van den Hondel CA, can den Broek HW. Cotransformation of Aspergillus nidulans: a tool for replacing fungal genes. Mol Genet Genomics. 1987;209:71–77.
- Kohli A, Twyman RM, Abranches R, Wegel E, Shaw P, Christou P, Stoger E. Transgene integration, organization and interaction in plants. Plant Mol Biol. 2003;52:247–258.
- Sakaguchi T, Amari S, Nagashio N, Murakami Y, Yokoyama K, Tamiya E. Genetic transformation of Trichosporon cutaneum with a plasmid, pAN 7–1, from filamentous fungi. Biotechnol Lett. 1998;20(9):851–855.
- Punt PJ, Oliver RP, Dingemanse MA, Pouwels PH, van den Hondel CAMJJ. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene. 1987;56:117–123.
- Nakayashiki H, Kiyotomi K, Tosa Y, Mayama S. Transposition of the retrotransposon MAGGY in heterologous species of filamentous fungi. Genetics. 1999;15:693–703.
