Abstract
The complete mRNA sequence of watermelon Rab18 gene was amplified through the rapid amplification of cDNA ends (RACE) method. The full-length mRNA was 1010 bp containing a 645 bp open reading frame, which encodes a protein of 214 amino acids. Sequence analysis revealed that watermelon Rab18 protein shares high homology with the Rab18 of cucumber (99%), muskmelon (98%), Morus notabilis (90%), tomato (89%), wine grape (89%) and potato (88%). Phylogenetic analysis revealed that watermelon Rab18 gene has a closer genetic relationship with Rab18 gene of cucumber and muskmelon. Tissue expression profile analysis indicated that watermelon Rab18 gene was highly expressed in root, stem and leaf, moderately expressed in flower and weakly expressed in fruit.
Introduction
Rab GTPase family 18 (Rab18) is a member of Rab GTPases. Most Rab GTPases contain a lipid modification site at the C-terminus, with sequence motifs CC, CXC or CCX. Lipid binding is essential for membrane attachment, a key feature of most Rab proteins.[Citation1–3] Mammalian Rab18 is implicated in endocytic transport and is expressed most highly in polarized epithelial cells. In human and mouse cells, Rab18 has been identified in lipid droplets, organelles that store neutral lipids.[Citation1–6] Recent research showed that the mutations in RAB18 gene are correlated to the Warburg micro syndrome and Martsolf syndrome.[Citation6,Citation7] In plants, Rab18 gene over-expression can improve the salt stress, cold stress, freezing stress and drought stress resistance in Arabidopsis thaliana,[Citation8–12] and is also related to Oryza sativa salinity stress response.[Citation13]
Based on the above-mentioned points, Rab18 gene is considered as an important gene for animals and plants. It has been identified in many plants such as cucumber and muskmelon. However, watermelon Rab18 gene has not been reported yet.
In the present paper, we isolated the full-length mRNA sequence of watermelon Rab18 gene and performed sequence analysis and tissue expression analysis. These will establish the primary foundation of utilizing watermelon Rab18 gene to improve watermelon production.
Materials and methods
Sample collection, RNA extraction and first-strand cDNA synthesis
Watermelon plants (Chinese cultivar Xinlvbao) were grown naturally with normal irrigation and fertilization. The tissues, including leaves, stem, root, flower and fruit (fruit of watermelon at immature white, white-pink flesh, red flesh and over-ripe stages), were harvested and immediately frozen in liquid nitrogen and stored at −80 °C. Total RNA extraction and first-strand cDNA synthesis for these tissue samples were performed as the methods described by Liu.[Citation14]
5′ and 3′-RACE
5′- and 3′-RACE (rapid amplification of cDNA ends) were performed using SMARTTM RACE cDNA Amplification Kit. For watermelon Rab18 gene, the gene-specific primers (GSPs) were designed based on one watermelon EST (expressed sequence tag) sequence: GD177891. 5′-RACE GSP: 5′-GCCAAAAGACTAGGCGTGTCCAATA-3′, 3′-RACE GSP: 5′-ATGGATGCCTATTTCTCGAATGCAG-3′.
RACE touchdown PCRs (polymerase chain reactions) were carried out by heating the reaction mixture as follows: five cycles of 94 °C for 30 s and 72 °C for 3 min, followed by five cycles of 94 °C for 30 s, 70 °C for 30 s and 72 °C for 3 min, and finally with 25 cycles of 94 °C for 30 s, 65 °C for 30 s and 72 °C for 3 min to terminate the reaction. These RACE PCR products were then cloned into PMD18-T vector (TaKaRa, China) and sequenced bidirectionally with the commercial fluorometric method.
Quantitative real-time PCR (qRT-PCR) for tissue expression profile analysis
Quantitative real-time PCR (qRT-PCR) for evaluating the level of mRNA for Rab18 gene was performed by the ABI Prism 7300 Sequence Detection Systems (Applied Biosystems, Foster City, CA, USA). A 25 µL of reaction mixture used for PCR reaction contained 1 µL SYBR Green real-time PCR Master Mix, 100 ng cDNA template and 200 nmol/L each primer. Conditions for real-time PCR were as follows: an initial denaturation at 95 °C for 3 min, 40 cycles of 95 °C for 15 s, optimal annealing temperature for each specific primer for 15 s () and 72 °C for 20 s. The gene relative expression levels were quantified relative to the expression of the reference gene, actin (GenBank accession no. GU565958), by employing the 2−ΔΔCt value model.[Citation15]
Table 1. qRT-PCR primers for watermelon Rab18, actin genes and annealing temperature.
Sequence analysis
Gene prediction of cDNA sequence was performed by GenScan software (http://genes.mit.edu/GENSCAN.html). Theoretical isoelectric point (pI) and molecular weight (Mw) of the deduced protein were computed using the Compute pI/Mw Tool (http://www.expasy.org/tools/pi_tool.html). Protein analysis was carried out using the BLAST tool at the National Center for Biotechnology Information (NCBI) server (http://www.ncbi.nlm.nih.gov/BLAST) and the Clustalw software (http://www.ebi.ac.uk/clustalw).
Results and discussion
RACE results for watermelon Rab18 gene
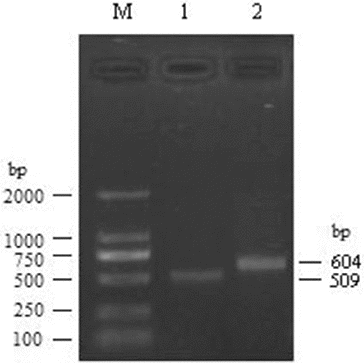
For watermelon Rab18 gene, through 5′-RACE, one PCR product of 604 bp was obtained. The 3′-RACE product was 509 bp. These products were then cloned to T-vector and sequenced. Taken together, a 1010 bp cDNA complete sequence was finally obtained ().
Sequence analysis
BLAST analysis of this cDNA nucleotide sequence revealed that the gene is not homologous to any of the known watermelon genes and it was then deposited into the Genbank database (accession number: KM235552). Results showed that this 1010 bp cDNA sequence represents one single gene which encodes 214 amino acids (). The theoretical pI) and Mw of the deduced protein of this watermelon gene were also computed. The pI of watermelon Rab18 is 5.97. The Mw of this putative protein is 23762.12 Da.
Figure 2. The complete mRNA of watermelon Rab18 gene and its encoding amino acids. * indicates the stop codon.

Further BLAST analysis of this protein revealed that watermelon Rab18 gene shares high homology with Rab18 gene of cucumber (accession number: XP_004146786, 99%), muskmelon (accession number: XP_008445776, 98%), Morus notabilis (accession number: EXB37658, 90%), tomato (accession number: XP_004237716, 89%), wine grape (accession number: XP_002267387, 89%) and potato (accession number: XP_006356316, 88%) (). Its conserved domain was identified as Rab18 ().
From the results obtained above, it can be concluded that this protein is watermelon Rab18, and this new gene is watermelon Rab18 gene.
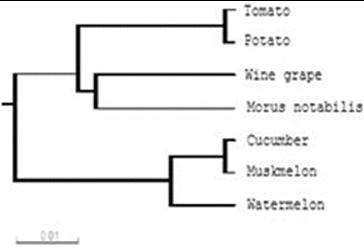
Based on the results of the alignment of different species of Rab18 proteins, a phylogenetic tree was constructed using the Clustalw software, as shown in . The phylogenetic analysis revealed that watermelon Rab18 gene has a closer genetic relationship with Rab18 gene of cucumber and muskmelon.
Tissue expression profile
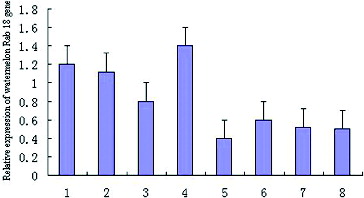
Tissue expression profile analysis was carried out and the results revealed that watermelon Rab18 gene was highly expressed in root, stem and leaf, moderately expressed in flower and weakly expressed in fruit ().
Figure 6. Expression analysis of Rab18 gene mRNA in various watermelon tissues: 1, leaves; 2, stem; 3, flower; 4, root; 5, fruit (immature white stage); 6, fruit (white-pink flesh stage); 7, fruit (red flesh stage); 8, fruit (over-ripe stage).
Modern bioinformatics has revealed that virtually all (99%) of the protein-coding genes in humans align with homologues in mouse, and over 80% are clear 1:1 orthologues for human and mouse both belong to mammalian.[Citation14,Citation16] This extensive conservation in protein-coding regions implied that this conservation of protein-coding sequences may be expected in watermelon and other plants of Cucurbitaceae. From the sequence analysis of Rab18 genes, it can be seen that the coding sequences of Rab18 genes were highly conserved in three Cucurbitaceae plants–watermelon, cucumber and muskmelon.
The phylogenetic tree analysis revealed that watermelon Rab18 gene has a closer genetic relationship with Rab18 gene of cucumber and muskmelon. This implied that we can use cucumber and muskmelon as model organisms to study watermelon Rab18 gene or use watermelon as a model organism to study Rab18 gene of cucumber and muskmelon.
From the tissue distribution analysis in our experiment, it can be seen that watermelon Rab18 gene was differentially expressed in some tissues. For plants, Rab18 gene expression is related to salt stress, cold stress, freezing stress, salinity stress and drought stress resistance [Citation1–13]; the suitable explanation for this under current conditions is that the biological activity of Rab18 was present diversely in different tissues.
Conclusions
We first isolated watermelon Rab18 gene and performed necessary sequence analysis and tissue expression profile analysis. These established the primary foundation of utilizing watermelon Rab18 gene to improve the production of watermelon and benefit humans in the future.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Seabra MC, Wasmeier C. Controlling the location and activation of Rab GTPases. Curr Opin Cell Biol. 2004;16:451–457.
- Dansako H, Hiramoto H, Ikeda M, Wakita T, Kato N. Rab18 is required for viral assembly of hepatitis C virus through trafficking of the core protein to lipid droplets. Virology. 2014;462–463C:166–174.
- Gerondopoulos A, Bastos RN, Yoshimura S, Anderson R, Carpanini S, Aligianis I, Handley MT, Barr FA. Rab18 and a Rab18 GEF complex are required for normal ER structure. J Cell Biol. 2014;205:707–720.
- Ozeki S, Cheng J, Tauchi-Sato K, Hatano N, Taniguchi H, Fujimoto T. Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane. J Cell Sci. 2005;118:2601–2611.
- Cheng CY, Yang AC, Huang CC, Liu ME, Liou YJ, Wu JC, Tsai SJ, Lin CP, Hong CJ. The association of RAB18 gene polymorphism (rs3765133) with cerebellar volume in healthy adults. Cerebellum. 2014; 13(5):616–622.
- Carpanini SM, McKie L, Thomson D, Wright AK, Gordon SL, Roche SL, Handley MT, Morrison H, Brownstein D, Wishart TM, Cousin MA, Gillingwater TH, Aligianis IA, Jackson IJ. A novel mouse model of Warburg micro syndrome reveals roles for RAB18 in eye development and organisation of the neuronal cytoskeleton. Dis Model Mech. 2014;7(6):711–722.
- Handley MT, Morris-Rosendahl DJ, Brown S, Macdonald F, Hardy C, Bem D, Carpanini SM, Borck G, Martorell L, Izzi C, Faravelli F, Accorsi P, Pinelli L, Basel-Vanagaite L, Peretz G, Abdel-Salam GM, Zaki MS, Jansen A, Mowat D, Glass I, Stewart H, Mancini G, Lederer D, Roscioli T, Giuliano F, Plomp AS, Rolfs A, Graham JM, Seemanova E, Poo P, García-Cazorla A, Edery P, Jackson IJ, Maher ER, Aligianis IA. Mutation spectrum in RAB3GAP1, RAB3GAP2, and RAB18 and genotype–phenotype correlations in Warburg micro syndrome and Martsolf syndrome. Hum Mutat. 2013;34(5):686–696.
- Geng Y, Wu R, Wee CW, Xie F, Wei X, Chan PM, Tham C, Duan L, Dinneny JR. A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell. 2013;25:2132–2154.
- Gosti F, Bertauche N, Vartanian N, Giraudat J. Abscisic acid-dependent and -independent regulation of gene expression by progressive drought in Arabidopsis thaliana. Mol Gen Genet. 1995;246:10–18.
- Puhakainen T, Hess MW, Mäkelä P, Svensson J, Heino P, Palva ET. Overexpression of multiple dehydrin genes enhances tolerance to freezing stress in Arabidopsis. Plant Mol Biol. 2004;54:743–753.
- Lång V, Palva ET. The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol Biol. 1992;20:951–962.
- Shi H, Chen L, Ye T, Liu X, Ding K, Chan Z. Modulation of auxin content in Arabidopsis confers improved drought stress resistance. Plant Physiol Biochem. 2014;82:209–217.
- Jha Y, Sablok G, Subbarao N, Sudhakar R, Fazil MH, Subramanian RB, Squartini A, Kumar S. Bacterial-induced expression of RAB18 protein in Oryza sativa salinity stress and insights into molecular interaction with GTP ligand. J Mol Recognit. 2014;27:521–527.
- Liu GY. Isolation, sequence identification and tissue expression profile of two novel soybean (Glycine max) genes – vestitone reductase and chalcone reductase. Mol Biol Rep. 2009;36(7):1991–1994.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25(4):402–408.
- Hardison RC. Comparative genomics. PLoS Biol. 2003;1:E58.