Abstract
The function of several families of clustered regularly interspaced palindrome repeats (CRISPRs) in prokaryotic genomes was recently found to be related to the protection of bacterial cells against the expression of foreign DNA, originating from plasmids or bacteriophages. The present study was the first attempt to screen a broader number of Lactobacillus delbrueckii ssp. bulgaricus strains, widely used in yoghurt and cheese production, for the presence of CRISPRs. Database search of four completely sequenced L. delbrueckii ssp. bulgaricus genomes indicated the presence of CRISPR2 in three of them - ATCC 11842, ATCC BAA-365 and ND02, and the presence of CRISPR3 in strain 2038. In the first three strains, the CRISPR2 was invariably located between a 3′–5′ exonuclease gene and a gene for a ppGpp-synthetase. The location of CRISPR3 in strain 2038 was between a histidine-kinase gene and an acetyl-CoA acetyltransferase gene, 2 kbp downstream of the CRISPR2 locus in ATCC 11842. Specific primers were designed to amplify with polymerase chain reaction the target regions containing the potential CRISPR2 and / or CRISPR3 in a total of 33 L. delbrueckii ssp. bulgaricus strains. Thirteen strains yielded a high molecular mass product corresponding in size and location to CRISPR2 of the type strain ATCC 11842, while another 17 strains indicated the presence of potential CRISPR3, analogous to that of strain 2038. Three strains did not indicate the presence of CRISPRs. Interestingly, none of the tested strains carried both CRISPR2 and CRISPR3 simultaneously in its genome at the investigated region.
Introduction
Bacteria from the Lactobacillus delbrueckii ssp. bulgaricus species are a basic component of yoghurt and white-brined cheese starters. Compared to the phage attack on Streptococcus thermophilus, phage infection on L. delbrueckii ssp. bulgaricus cultures occurs much more rarely.[Citation1,Citation2] Nevertheless, bacteriophages that attack the L. delbrueckii group of subspecies are well documented in the literature.[Citation3–6] Recently, the first local isolate of L. delbrueckii ssp. bulgaricus bacteriophage was classified into group ‘b’ L. delbrueckii bacteriophages, based on its partial genome sequencing.[Citation6]
The clustered regularly interspaced palindrome repeats (CRISPRs) were first described in the genome of Escherichia coli K-12 and their presence was confirmed for a multitude of bacterial and archaeal genomes.[Citation7,Citation8] Recently, these palindrome repeats and the cas-genes, associated with them, have been found to function as a new defence mechanism in prokaryotic cells against invading phages and plasmids.[Citation9,Citation10] The regular repeats are interspaced with short sequences named ‘spacers’ which derive from foreign genetic elements.[Citation9,Citation11,Citation12] When, for example, the bacterial host is under a bacteriophage attack, it acquires a new spacer sequence within its CRISPR locus that matches a DNA sequence in the phage genome, referred to as a ‘protospacer’.[Citation13,Citation14] The complete set of repeats and spacers is then transcribed and processed into individual crRNAs that interact with the viral DNA, facilitating its cleavage and rendering the bacterial cell immune to infection by the particular phage.[Citation14–16] Depending on the organization of the cas-genes and the sequence of the palindrome repeats, four classes of CRISPRs are described and numbered from 1 to 4. As an example, in S. thermophilus, all four classes of CRISPR/cas systems are described as CRISPR1 and CRISPR3, which are considered to be the most active ones.[Citation14,Citation16]
As data on CRISPRs in the genome of L. delbrueckii sp. bulgaricus are still lacking, in the present study we reported the results from the database search of potential CRISPRs in this subspecies and from the screening of 33 strains for the presence of potential CRISPR2 and CRISPR3 loci in their genome.
Materials and methods
Bacterial strains, culture conditions and DNA isolation
Thirty-three L. delbrueckii ssp. bulgaricus strains maintained in the LBB culture collection (LB Bulgaricum PLC, Sofia, Bulgaria) were included in the study. All cultures were grown in MRS medium (peptone – 10 g/L; meat extract – 8 g/L; yeast extract – 4 g/L; glucose – 20 g/L; sodium acetate trihydrate – 5 g/L; Tween-80 – 1 g/L; dipotassium hydrogen phosphate – 2 g/L; triammonium citrate – 2 g/L; magnesium sulfate heptahydrate – 0.2 g/L; manganese sulfate – 0.05 g/L; pH was adjusted to 6.2) [Citation17] for 24 hours at 37 °C. We used 5 mL of the cultures for DNA isolation with the GenElute Bacterial Genomic DNA Kit (Sigma-Aldrich, St. Louis, MO, USA), according to the manufacturer's instructions. A list of the tested L. delbrueckii ssp. bulgaricus strains is presented in .
Table 1. List of the tested bacterial strains with the presence (+) or absence (−) of the respective products from a CRISPR2-specific and CRISPR3-specific PCR amplification.
Database search for CRISPRs and primer design
Four publicly available genomes of L. delbrueckii ssp. bulgaricus' strains ATCC 11842, ATCC BAA-365, ND02 and 2038 (GenBank Acc. Nos. NC_008054; NC_008529; NC_014727 and NC_017469) were searched for CRISPRs with the CRISPRFinder software.[Citation18] Conservative regions upstream and downstream of CRISPR2 and CRISPR3 were identified by alignment with the CLC Sequence Viewer (www.clcbio.com) and suitable primers were designed with the PrimerBLAST tool.[Citation19] A complete list of the primers, their sequence and location is presented in .
Table 2. List of the designed primers and their location in the analysed genomic region.
Polymerase chain reaction (PCR) amplifications
All amplifications were performed on a 9600 GeneAmp PCR System (Perkin-Elmer, Norwalk, Connecticut) in a 25 μL reaction mixture consisting of diluted VWR Taq DNA Polymerase Master Mix (VWR International, Haasrode, Belgium), 50 ng template DNA and 10 pmol of each primer. The PCR programme was as follows: one cycle of 3 min at 95 °C, 30 cycles of 30 s at 93 °C, 30 s at 60 °C and 1 min at 72 °C, and one cycle of 7 min at 72 °C. The potential CRISPR2 and CRISPR3 regions were amplified with primer pairs pr21 / pr22 and pr35 / pr32, respectively. Additionally, primers pr31 / pr32 were used to amplify a short region to confirm the absence of CRISPR3 in the searched location. The position of the primers within the analysed region is illustrated in . The amplified products were separated by electrophoresis in a 2% agarose gel in Tris-Acetate-EDTA buffer (40 mmol/L Tris-acetate and 1 mmol/L EDTA, pH 8.3) at 100 V and visualized after staining with ethidium bromide. The size of the obtained product was determined using a suitable size marker (Gene Ruler 100 bp Plus DNA Ladder, Thermo Scientific, Pittsburgh, PA, USA).
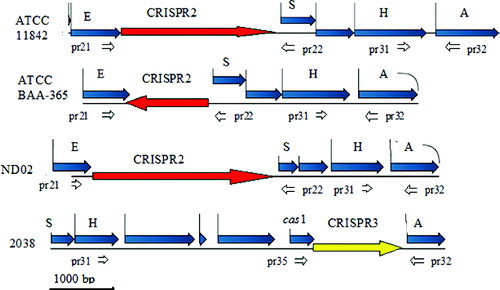
Figure 1. Schematic representation of the location of CRISPR2 and CRISPR3 in the genomes of Lactobacillus delbrueckii ssp. bulgaricus strains ATCC 11842, ATCC BAA-365, ND02 and 2038 and the position of the designed primers.
Note: Acetyl-CoA acetyltransferase gene (A); 3′–5′ exonuclease gene (E); histidine-kinase gene (H); ppGpp-synthetase gene (S); CRISPR3-associated gene (cas1); specific primers (pr21 to pr35).
Results and discussion
The database search of the four completely sequenced L. delbrueckii ssp. bulgaricus genomes indicated the presence of CRISPR2 in three of them – ATCC 11842, ATCC BAA-365 and ND02 and the presence of CRISPR3 in strain 2038.
In the first three strains the CRISPR2 was invariably located between a 3′–5′ exonuclease gene and a gene for a ppGpp-synthetase (). In strain 2038, the region preceding the analogue of the ppGpp-synthetase gene contained completely different genes instead of CRISPR2. The location of CRISPR3 of strain 2038 was between a histidine-kinase gene and an acetyl-CoA acetyltransferase gene, 2 kbp downstream of the CRISPR2 locus in ATCC 11842 (). In strains ATCC 11842, ATCC BAA-365 and ND02, which did not have the CRISPR3 region, the acetyl-CoA acetyltransferase gene followed immediately after the histidine-kinase gene. The CRISPRFinder software listed 40, 20 and 64 spacers within CRISPR2 of strains ATCC 11842, ATCC BAA-365 and ND02, respectively, and 19 spacers within CRISPR3 of strain 2038. A list of the spacers within CRISPR2 and CRISPR3 of strains ATCC 11842 and 2038 is presented in .
Table 3. List of spacers within CRISPR2 and CRISPR3 of Lactobacillus delbrueckii ssp. bulgaricus strains ATCC 11842 and 2038, respectively, retrieved by CRISPRFinder software.[Citation18]
Using the specific primers pr21/pr22 for the CRISPR2 region, 13 strains yielded a high molecular mass product, corresponding in size (>3 kbp) and location to CRISPR2 of the type strain ATCC 11842 (). The separation of the CRISPR2-specific amplified product for strain ATCC 11842 and five representative ‘CRISPR2-positive’ strains is presented in (A). Neither of the ‘CRISPR2-positive’ strains yielded an amplification product with the CRISPR3-specific primer pair pr35/pr32 (, (A)). Additionally, for all ‘CRISPR2-positive’ strains a short amplification product was obtained with primers pr31 / pr32 that proved that the histidine-kinase gene was followed immediately by the acetyl-CoA acetyltransferase gene with no insertion of CRISPR3 between them. A visualization of this short amplification product for strain ATCC 11842 and five representative ‘CRISPR2-positive’ strains is included in (A).
Figure 2. Analysis of the presence/absence of CRISPR2 (primer pair pr21/pr22; lanes 1–6 and 19–24), CRISPR3 (primer pair pr35/pr32; lanes 7–12 and 25–30) and histidine-kinase / acetyl-CoA acetyltransferase gene (primer pair pr31/pr32; lanes 13–18 and 31–36) specific amplification products in a selection of ‘CRISPR2-positive’ (A) or ‘CRISPR3-positive’ (B) L. delbrueckii ssp. bulgaricus strains.
Note: representative ‘CRISPR2-positive’ strains: ATCC 11842 (lanes 1, 7, 13), B5/1S (2, 8, 14), B60/8S (3, 9, 15), B208/5S (4, 10, 16), B226/2S (5, 11, 17) and B278/2S (6, 12, 18) (A); representative ‘CRISPR3-positive’ strains: B37/5S (19, 25, 31), B54/6J (20, 26, 32), B56/6J (21, 27, 33), B69/3J (22, 28, 34), B127/8S (23, 29, 35) and B215/6S (24, 30, 36) (B). 100 bp Plus DNA ladder (M).
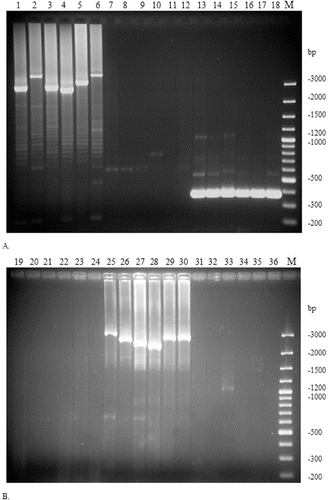
Another 17 strains gave positive amplification with primers pr35/pr32, specific for the CRISPR3 region of strain 2038, indicating the presence of potential CRISPR3 in these cultures (). The separation of the CRISPR3-specific amplified product for strain 2038 and five representative ‘CRISPR3-positive’ strains is presented in (B). All of the ‘CRISPR3-positive’ strains failed to produce amplification product with the CRISPR2-specific primer pair pr21/pr22 suggesting the absence of CRISPR2 in these strains (). The ‘CRISPR3-positive’ strains also did not yield the short amplification product of primer pairs pr31/pr32, possibly due to the large size of the targeted region, resulting from the insertion of CRISPR3 between the histidine-kinase and acetyl-CoA acetyltransferase genes. The absence of CRISPR2-specific products and the short amplification product of primers pr31/pr32 for strain 2038 and five representative ‘CRISPR3-positive’ strains is demonstrated in (B).
The indirect evidence that the amplified region contains CRISPRs was the formation of amplification sub-products with regularly increasing size, which formed a typical ladder-like electrophoretic pattern ((A) and 2(B)). This phenomenon can be explained by the formation of loops in some of the template molecules due to internal annealing of the palindromic repeats that prematurely terminate the elongation process. Interestingly, none of the tested strains carried both CRISPR2 and CRISPR3 simultaneously at the investigated region of their genome. Three strains did not indicate the presence of CRISPRs, due to their absence in the genome of the strains, or a completely different location of the potential CRISPRs. A complete list of all strains with the respective amplification products is given in .
Conclusions
Thirty-three strains of L. delbrueckii ssp. bulgaricus were analysed for the presence of CRISPR2 and CRISPR3 in their genomes. For 13 and 17 of them it was found that CRISPR2 and CRISPR3, respectively, were present and had the same location as that in strains ATCC 11842 and 2038. Neither strain contained the two CRISPRs simultaneously. Although the tested strains may still have CRISPRs in different locations that were not analysed in this study, it seems an interesting finding that with respect to the presence and location of CRISPR2 and CRISPR3 L. delbrueckii ssp. bulgaricus strains could be divided into two lineages - that of strain ATCC 11842 and that of strain 2038. The actual work on determining the sequences of spacers within the CRISPRs of L. delbrueckii ssp. bulgaricus strains is still ahead and may reveal important evolutionary relations between different cultures and highlight the history of particular strains with respect to encounters with foreign DNA.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Ishlimova D, Urshev Z, Stoyancheva G, Petrova P, Minkova S, Doumanova L. Genetic diversity of bacteriophages highly specific for Streptococcus thermophilus strain LBB.A. Biotechnol Biotechnol Equipment. 2009;23:1340–1345.
- Ishlimova D, Urshev Z, Alexandrov M, Doumanova L. Diversity of bacteriophages infecting the Streptococcus thermophilus component of industrial yogurt starters in Bulgaria. In: Galabov A, Nikolaeva-Glomb L, editors. Third congress of virology. Proceedings and Abstracts; 2012 October 25–27; Sofia, Bulgaria: The Stefan Angeloff Institute of Microbiology, Bulgarian Academy of Sciences; 2012.
- Chow J, Batt C, Sinskey A. Characterization of Lactobacillus bulgaricus bacteriophage ch2. Appl Environ Microbiol. 1988;54:1138–1142.
- Mata M, Trautwetter A, Luthaud G, Ritzenthaler P. Thirteen virulent and temperate bacteriophages of Lactobacillus bulgaricus and Lactobacillus lactis belong to a single DNA homology group. Appl Environ Microbiol. 1986;52:812–818.
- Vasala A, Dupont L, Baumann M, Ritzenthaler P, Alatossava T. Molecular comparison of the structural proteins encoding gene clusters of two related Lactobacillus delbrueckii bacteriophages. J Virol. 1993;67:3061–3068.
- Aleksandrova V, Ishlimova D, Urshev Z. Classification of Lactobacillus delbrueckii ssp. bulgaricus phage Gb1 into group ‘b’ Lactobacillus delbrueckii bacteriophages based on its partial genome sequencing. Bulg J Agric Sci. 2013;19(2):90–93.
- Nakata A, Amemura M, Makino K. Unusual nucleotide arrangement with repeated sequences in the Escherichia coli K-12 chromosome. J Bacteriol. 1989;171:3553–3556.
- Jansen R, van Embden J, Gaastra W, Schouls L. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–1575.
- Bolotin A, Quinquis B, Sorokin A, Erlich D. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561.
- Makarova K, Grishin N, Shabalina S, Wolf Y, Koonin E. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:1–26.
- Mojica F, Díez-Villaseñor C, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182.
- Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663.
- Deveau H, Garneau J, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Ann Rev Microbiol. 2010;64:475–493.
- Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170.
- Garneau J, Dupuis M-É, Villion M, Romero D, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán A, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71.
- Ishlimova D, Urshev Z. Evolution of CRISPR1 and CRISPR3 in spontaneous phage-resistant mutants of Streptococcus thermophilus strain LBB.A. Biotechnol Biotechnol Equipment. 2013;27:3966–3971.
- de Man J, Rogosa M, Sharpe M. A medium for the cultivation of lactobacilli. J Appl Bact. 1960;23:130–135.
- Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucl Acids Res. 2007;35:W52–W57.
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden T. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012;13:134–144.
