?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
The aim of the present study was to establish a colony lift assay for medullary thyroid carcinoma (MTC) cells and use it to screen a single-chain variable fragment antibody phage library targeted against MTC. This library was enriched with ‘adsorption–elution–amplification' in several selection rounds (SRs) and was then tested by a colony lift assay and random selection in each SR. The positive clones were scored among assay clones and the proportions that were obtained by each approach were compared. The results showed that positive clones for MTC antigen across one to several SRs accounted, respectively, for 0% (0/96), 3.13% (3/96), 10.42% (10/96) and 58.33% (56/96) in the random pick/selection approach, and 0.63% (2/318), 6.06% (12/198), 12.35% (20/162) and 63.51% (94/148) in the colony lift assay (SR1, χ2 = 0.607, P = 0.436; SR2, χ2 = 1.151, P = 0.283; SR3, χ2 = 0.218, P = 0.640; SR4, χ2 = 0.660, P = 0.417). There was no significant difference between the two methods, which suggested that the rates of positive clone selection by colony lift assay were consistent with those of random pick. However, the former method can test many more positive clones simultaneously. Thus, the established new colony lift assay method for panning MTC single-chain variable fragment (scFv) library could be considered effective, simple to manipulate and design, robust and convenient in screening a scFv antibody phage library.
Abbreviations
| BSA: | = | bovine serum albumin |
| HRP: | = | horseradish peroxidase |
| IPTG: | = | isopropyl-β-D-thiogalactoside |
| MTC: | = | medullary thyroid carcinoma |
| PBS: | = | phosphate buffered saline |
| PEG: | = | polyethylene glycol |
| scFv: | = | single-chain variable fragment |
| SR: | = | selection round |
| PRU: | = | protein recognition units |
| DAB: | = | diaminobenzidine |
| HAMA: | = | human anti-mouse antibodies |
Introduction
After synthesizing an anti-tumour single-chain variable fragment (scFv) antibody phage library, it is vital to create a reasonable, effective and feasible selection strategy. Anti-tumour scFv antibody library screening is designed according to the principle of an antigen and antibody specificity combination. It is used to show the characteristics of the phage antibody that complexes with the antigen.[Citation1]
The currently established screening method is ‘adsorption–elution–amplification,’ which is a process of randomly picking individual clones and testing them in sequence ‘one-by-one’ for cell binding. However, one major drawback of this strategy is the fact that clones that are present at a low frequency can hardly be recovered. Oftentimes, more than two or three SRs can increase the possibility of recovering positive clones, although this might lead to a significant bias towards a few dominant clones with obvious growth advantages. There are also considerations such as a certain degree of ‘blindness’ inherent in the procedures, a large workload and low efficiency.
By contrast, the colony lift assay [Citation2–4] enables prompt and efficient phage library screening for clones binding to cells, and it enables detection and isolation of phage antibody clones that are present at low frequency. The essence of the colony lift assay lies in the fact that phage-infected bacteria will produce soluble scFvs if plated on isopropyl-β-D-thiogalactoside (IPTG)-containing agar plates. The released scFv will diffuse through the bacterial filter and will bind to the antigen if it is immune cross-reactive to it, and then the antigen-coated filter is placed under the bacterial filter.
The scFv bound to the antigen-coated filter can be detected by staining the antigen-coated filter, using antibodies specific for the tag fused to the scFv. Thus, bacterial colonies producing scFv of determined specificity can be identified by this method, allowing picking of colonies infected with antigen specific, ‘binder’ phages. This ‘directed’ picking by the colony lift assay has a clear advantage in random picking bacterial colonies, especially in the context of phage libraries with a low frequency of binding clones. Up to now, to the best of our knowledge, this approach has not been reportedly used for medullary thyroid carcinoma (MTC) scFv antibody phage library screening. Thus, the aim of the present study was to develop a modified colony lift assay with cell-coated filters in an attempt to use this method in setting up a large capacity of scFv phage antibodies targeted against MTC at the beginning of a large-scale screen. In addition, an attempt was made to fine screen the library in the next step with the intention of identifying a large number of positive bacteria.
Materials and methods
Main materials and reagents
RPMI-1640 medium (Gibco), M13K07 helper phage (Pharmacia Amersham), Escherichia coli TG1 (NEB), HRP∕anti-E-Tag conjugate (Abcam; clone number – ab19400), bovine serum albumin (BSA) (Amresco), polyethylene glycol (PEG) (Sigma), IPTG (Merck), Tween-20 (Amresco) and glutaraldehyde solution (Sigma) were used. The bacterial culture medium used in our studies was double-strength YT medium. We also used double-strength YT-AG culture plates that were supplemented with 100 μg/mL ampicillin and 5% glucose (also referred to as AG plates). The master plates used in our study contained double-strength YT-AI plates that were supplemented with 100 μg/mL ampicillin and 0.1 mmol/L IPTG (also referred to as AI plates).
The MTC cells used in the experiments were originally isolated from five patient specimens that were pathologically diagnosed after surgery at our hospital.
Single-chain variable fragment phage antibodies against medullary thyroid carcinoma (MTC scFv) were constructed and stored in our laboratory. Nitrocellulose membranes (PALL P-N664865) were used for the method of coating of MTC cells onto nitrocellulose filters to obtain an MTC-coated filter.
Cell culture
The MTC cells were cultured in RPMI-1640 medium that was supplemented with 10% foetal calf serum plus 1% penicillin and streptomycin, and cultured at 37 °C and under a 5% CO2 atmosphere in air. When cells had grown to a near-monolayer confluence, we subcultured the cells or collected them aside for further use.
Coating MTC cells onto filters
The MTC cells that were intended to be coated onto nitrocellulose filters were washed and re-suspended in RPMI-1640 medium at a density of 1 × 106 cells/mL. Six millilitres of cell suspension was pipetted onto a 90-mm nitrocellulose filter (pore size of 0.2 μm) and placed in a 10-cm diameter Petri dish. The dish was then incubated for at least 4 h at room temperature. The supernatant was removed, and the cells that remained non-coated were counted to determine the coating efficiency (repeated three times). The cell-coated filter was washed with PBS and blocked with 10 mL PBS/4% low-fat dried milk proteins for 1 h at room temperature. The blocked filter was washed twice with PBS and air-dried for use.
Preparing phage scFv
A 200 μL MTC scFv bacterial suspension was added to 10 mL of AG medium and grown at 37 °C while shaking at 200 r/min until the culture had reached the log phase of growth. Next, we added 1 × 1011 PRU M13K07 helper phage to the cultured bacteria and incubated them at 37 °C for 1 h while shaking at 200 r/min. The bacteria were centrifuged for 10 min at 1000 × g, following which, the pellet was re-suspended in 10 mL of double-strength YT-AK medium containing 100 μg/mL ampicillin and 25 μg/mL kanamycin as selection antibiotics (further referred to as AK medium), which was then incubated overnight at 37 °C while shaking at 200 r/min. The overnight culture was centrifuged for 20 min at 1000 × g at 4 °C. The phage-containing supernatant was transferred to a new tube and precipitated with 1/5 volume (about 2 mL) of ice-cold PEG/NaCl for 1 h on ice. The phage suspension was centrifuged for 20 min at 10,000 × g at 4 ° C. The pellet was re-suspended in 2 mL ice-cold PEG/NaCl and 8 mL ice-cold PBS. After incubating for 30 min on ice, the suspension was centrifuged for 20 min at 10,000 × g at 4 °C. The pellet was re-suspended in 1 mL PBS/1% BSA, transferred to an Eppendorf tube®, and centrifuged for 1 min to remove debris. The supernatant containing phage scFv was collected and stored at 4 °C until the next SR.
Affinity enrichment of MTC scFv
Adsorption and elution of MTC scFv
A 6 mL MTC cell suspension was prepared and added in a 50-mL culture flask; then it was cultured at 37 °C under an atmosphere of 5% CO2 in air for 24 h. The culture medium was aspirated and the remaining cells were washed with PBS/1% BSA at 4 °C; then 5 mL of 0.05% glutaraldehyde solution was added and the culture was placed at 4 °C for 15 min. The glutaraldehyde solution was discarded and the cultured cells were washed three times with PBS. Then, the culture bottle or Petri dishes were filled with PBS/3% BSA, and incubated closed overnight at 4 °C. The solution was discarded, phage svFv (described above) were added at 4 mL and incubated at room temperature for 2 h. The phage scFv were removed and the preparations were washed six times. Each washing step was performed for 5 min with 4 mL PBS/0.05% Tween-20. After a final wash, 300 μL PBS/0.05% Tween-20 and 450 μL PBS/76 mmol/L citric acid (pH 2.5) were added and incubated for 10 min with shaking to elute bound phages. Elute phages were neutralized by adding 250 μL of 1 mol/L Tris-HCL (pH 7.4) and were allowed to infect E. coli TG1 bacteria to prepare a restricted (selected) library. The selection protocol was repeated three times, resulting in four sequential batches of selected library (first to fourth SRs).
Infection and amplification of E. coli TG1 bacteria with eluted phage
We added 100 μL E. coli TG1 bacterial suspension to 10 mL YT medium and cultured it at 37 °C while shaking at 200 r/min until the log phase. A phage infection was performed by mixing 3 mL log phage bacteria with 3 mL YT medium and above 1 mL eluted phages and incubating for 1 h at 37 °C while shaking at 200 r/min. The bacterial suspension was centrifuged for 15 min at 2000 × g at 4 °C and the pellet was re-suspended in 1 mL YT medium. A 250 μL bacterial suspension was plated on a 150-mm YT plate (master plate). Serial dilutions were then created from the bacterial suspension and plated on a 100-mm AT plate to titre the eluted phage (titration plates). The plates were then incubated overnight at 37 °C. The bacteria from the master plate were collected by scraping into 2 mL YT medium. Part of the bacterial suspension was used for detection and affinity enrichment for the next SR, and part of it was stored by adding 10% glycerol and freezing at −70 °C.
Colony lift assay
We used the method of coating MTC cells onto nitrocellulose filters to make an MTC-coated filter. We put the MTC-coated filter onto the surface of a 90-mm AI plate with the coated surface upward. The above phage-infected E. coli TG1 were plated in different dilutions on 90-mm AG plates and grown overnight at 37 °C. The plate containing single bacterial colonies was used for the colony lift assay (and this preparation is referred to as the original bacterial plate). Then, another non-coated filter was plated on top of the original bacterial plate (referred to as master filter). The master filter was removed from the original plate and placed with colonies facing upward on the cell-coated filter on the AI plate. Both filters were marked simultaneously with a needle for orientation purposes. Next, the AI plate was incubated overnight at 37 °C. The master filter was removed and placed onto a fresh AG plate with colonies facing upward, stored at 4 °C, and later used for picking selected clones. The cell-coated filter was removed from the AI plate, washed with 10 mL PBS/0.05% Tween-20 for 1 h while shaking, blocked in PBS/4% low-fat dried milk proteins for 1 h at room temperature and rinsed twice with PBS/0.05% Tween-20, followed by incubation with 10 mL of optimally diluted (1:5000) HRP/anti-E-Tag/PBS/ 4% low-fat dried milk proteins for 1 h at room temperature. The filter was washed with 10 mL PBS/0.05% Tween-20 for 30 min while shaking and then rinsed twice with PBS. We added 5 mL of H2O2 activated diaminobenzidine (DAB) solution to the filter until spots were visible. Washing the filter with water stopped the reaction. The spots from the cell-coated filter were used to identify the corresponding colonies on the original bacterial plate or on the master filter, i.e., the colonies that were responsible for producing specific scFv (see and ).
Figure 1. Principle of the colony lift assay. A cell-coated filter was placed on an IPTG-containing plate (AI plate). Bacteria infected with the selected phage library were plated on the AG plate, and a replica of the bacterial filter (master filter) was made and placed on top of the cell-coated filter. Upon overnight incubation, the master filter was removed, placed on a fresh AG plate and stored for further use. The cell-coated filter was stained with scFv-tag specific antibody coupled to HRP and developed using H2O2 activated DAB. Subsequently, colonies corresponding to obtained spots were identified on the master filter.
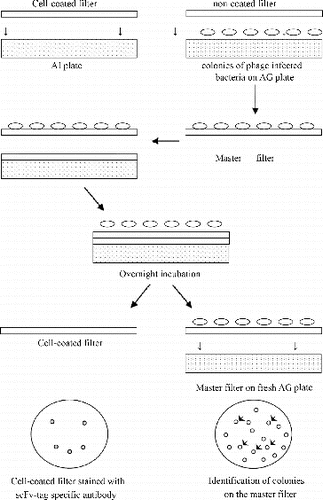
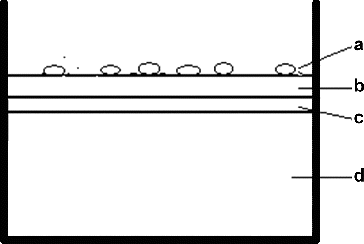
Figure 2. Simple principle of the colony lift assay: (A) bacterial clones from MTC scFv library; (B) master filter; (C) MTC-coated filter and (D) double strength YT + Amp + IPTG + agar.
Random picking detection
We randomly picked 96 bacterial clones from each master filter selection round, respectively, after preparing MTC phage scFv. This kind of MTC cell was simultaneously incubated in a 96-well culture plate, each well containing 1 × 106 cells, and cultured at 37 °C under an atmosphere of 5% CO2 in air for 24 h. The nutrient solution from each well was discarded and the wells were washed with PBS. Then, 20 µL 0.05% glutaraldehyde solution was added in each well to fix cells, after which they were incubated at 4 °C for 15 min and were washed three times with PBS. To each well, 200 μL of PBS/4% low-fat dried milk proteins were added and the samples were incubated for 1 h at room temperature and then rinsed twice with PBS. The liquid was discarded and the preparations were allowed to air dry. The MTC scFv preparation was then added, respectively, to each well (100 μL/well) and the M13K07 helper phage was used instead of phage antibodies as a negative control. Additionally, double-strength YT medium instead of phage antibody was used as a blank control. Following incubation for 2 h at 37 °C, the scFv solution was removed and each preparation was washed three times with PBS/0.05% Tween-20, the liquid was discarded and the preparation was air-dried. Then, optimally diluted (1:5000) HRP/anti-E-Tag/PBS /4% low-fat dried milk proteins were respectively added to each well (100 μL/well), which were then placed at 37 °C for 1 h. The solution was aspirated from each well, the wells were rinsed three times with PBS/0.05% Tween-20 and the liquid was removed. To each well, we added 100 μL H2O2 activated DAB solution at room temperature away from light and the absorption was measured at 405 nm (A405) with a standard spectrophotometer.
The following calculations were made:
where AE is the experimental group A405 value, AB is the blank control group A405 value and AN is the negative control group A405 value. A ratio greater than or equal to 2 was considered a positive result, and a ratio of less than 2 was considered as a negative result.
Statistical analysis
Statistical analysis was performed using SPSS 20.0 software. Measurement data were analysed using t-test; cell counts, by chi-square test and the results for the experiment parameters, using repeated measures analysis of variance. The threshold level of alpha was 0.05, and P-values less than or equal to 0.05 for differences were considered statistically significant.
Results and discussion
There is interest in finding new tumour antibodies to develop effective methods for early tumour diagnosis and immunotherapy, and the establishment of phage display technology may help to achieve this goal.[Citation5] The emergence of phage display technology and genetic engineering antibody library technology is a milestone, since single fundamental changes have occurred in resistance preparation methods. This technology is distinct from the rat source HAMA (human anti-mouse antibodies) single resistance response (mouse immune response) and includes an antibody diversity mechanism, the antibody variable region gene of VH and VL by RT-PCR cloning and amplification, and random combination electroporated into expression vector, making it possible to have humanized antibodies.[Citation6] Phage display technology links the phenotype, genotype, antibody antigen recognition and phage amplification ability together, creating an efficient expression and screening system.[Citation7,Citation8] In recent years, this technology has been used successfully to study cancers of the lung, the stomach, the liver, the colon and the head and neck, and it has been used to create an scFv antibody phage library. Here we present a modified colony lift assay with cell-coated filters for screening of scFv antibody phage library targeted against medullary thyroid carcinoma.
Efficiency of coating of nitrocellulose membrane filter
As a first step, we determined the efficiency of coating of the nitrocellulose membrane filter. After the coating, the non-coated cells that remained were approximately 0.87 × 106–1.26 × 106/mL, and they represented those that remained in the solution that was removed after coating. The coating contained about 4.74 × 106–5.13 × 106 cells per 90 mm2 filter, and the coating efficiency was around 79.0%–85.5%, indicating that it could be applied in the following experiments.
Colony lift assay on MTC cells using cell-coated filter
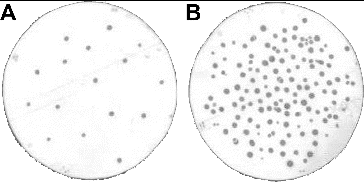
shows the result of the colony lift assay on MTC cells using the third SR, and scFv binding spots could be visualized on the cell-coated filter (). Corresponding colonies, responsible for the production of these cell-specific scFv antibodies (colony lift-positive clones), could be identified among all colonies on the master filter ((B)).
Comparison of efficiency of random picking method and colony lift assay
The positive clones for MTC antigen in the first to fourth SRs accounted for, respectively, 0% (0/96), 3.13% (3/96), 10.42% (10/96) and 58.33% (56/96) for randomly picked colonies, and 0.63% (2/318), 6.06% (12/198), 12.35% (20/162) and 63.51% (94/148) for the colony lift assay (SR1, χ2 = 0.607, P = 0.436; SR2, χ2 = 1.151, P = 0.283; SR3, χ2 = 0.218, P = 0.640; SR4, χ2 = 0.660, P = 0.417). A complete summary of these data is presented in . The results showed no significant differences between the two methods, suggesting that the rates of positive clones by the colony lift assay were comparable to those achievable by the random picking method. However, the major advantage of the colony lift assay compared to the random picking method is the fact that the colony lift assay can screen large numbers of clones simultaneously. We demonstrated that the colony lift assay was an effective, simple and convenient method for screening an scFv antibody phage library.
Table 1. Frequency of positive clones screened from MTC scFv library by random pick and colony lift assay.
Generally, there have been established several phage scFv library screening methods. Investigators have screened some meaningful tumour-specific antibodies, but screening with tumour-specific antibody currently remains infrequent.[Citation9] Existing screening methods are mainly divided into the following categories. (1) Screening of intact cells,[Citation10] directly with the expression of the target antigen cells to adsorb phage antibody, with enrichment of several rounds of affinity antibody libraries. Then, the last round of the enriched clones of bacteria can be randomly picked with the aim of testing them sequentially one by one. This method is applicable to cell-surface specific antibody screening,[Citation11] but some reports suggest that the screening results method may be unpredictable.[Citation12] (2) Liquid antigen screening,[Citation13] soluble resistance factors combined with phage antibody, original and combined with the support of antibodies in the packaging phage. This method can better retain the natural conformation of the antigen. (3) Group selection,[Citation14] by which it may be in some cases difficult to obtain a single-cell tissue specificity of antibody screening. (4) Using membrane protein or peptide molecules as antigens,[Citation15] which includes other processes similar to the aforementioned methods. These screening methods need to make several rounds of enrichment to ensure the removal of negative antibody library clones and increase the relative proportion of the positive clones. However, the disadvantage of this approach is that the enrichment process can cause loss of low-affinity clones, which might actually be highly significant and specific positive clones.
The detection method of randomly picking the clones does not easily detect the low frequency of rare clones, and picking out such clones sequentially one by one to extract the negative clone is incredibly labour-intensive, with concordantly high inspection costs and low efficiency that are inherent in the random selection procedure.[Citation16] Radosević et al. [Citation4] developed a colony lift assay with a cell-coated filter in 2003, and this method overcomes the above methods' shortcomings. Its main principle is that phage-infected bacteria will produce soluble scFv antibodies when plated on IPTG-containing agar plates and incubated overnight at 37 °C. These antibodies will diffuse through the bacterial filter and, if they are antigen-specific, will immunologically cross-react and bind to an antigen-coated filter placed beneath the bacterial filter. The filter-bound scFv can be detected using antibodies specific for scFv-tag. Detected scFv spots are used to identify bacterial colonies responsible for specific scFv production. These colonies can be picked and used for further phage antibody analysis. This method is essentially a primary screening method, and investigators can use this method to screen positive colonies for antibody libraries in the first instance, and then screen the positive colonies for further identification. This is produced by the combination of an scFv drop degree and an scFv gene sequence analysis.
We applied this method in our experimental systems to screen scFv phage antibodies against MTC cells.[Citation17] The results showed that cells were effectively coated on the nitrocellulose membrane filter. We used a colony lift assay with the membrane to detect the scFv library after enrichment, and in each selected round, we detected positive clones. In our experiment, a 9-cm agar dish with at least hundreds of scFv colonies could be successfully applied in a colony lift assay with a cell-coated filter. The frequency of positive clones was quite similar for both methods for the same SR. In addition, the frequency of positive clones increased upon subsequent SRs; however, the random picking method did not detect positive clones in the first SR. These results show that the colony lift assay with a cell-coated filter is theoretically consistent with the random picking method, demonstrating that the colony lift assay was quite efficient. The colony lift assay can also detect positive clones in the first SR easily and quickly, and only under conditions when positive clones were identified, which saves both time and unnecessary scoring of negative clones by the investigator. However, the random picking method was needed to detect each chosen clone from each SR to identify positive clones. This method is laborious and should generally be carried out three or four rounds after enrichment. However, enrichment processes were needed for the ‘adsorption–elution–amplification' process of multiple links and could easily cause low frequency and/or loss of low-affinity positive clones.
Thus, the random picking method is difficult for screening of such positive clones. By contrast, the colony lift assay also allows large numbers of clones to be screened simultaneously and can detect positive clones in the first SR following the enrichment. To some extent, the colony lift assay can overcome some of the identified drawbacks of the random picking method.
Conclusions
In conclusion, colony lift assay with cell-coated filter can be used in MTC phage scFv library screening, which can avoid the loss of low affinity of cloning in the process of enrichment, and these cloning could be significant specifically positive clones. Compared to randomly pick method which is not easy to detect the low frequency of rare cloning, and also one by one to pick out the negative clone to detect, the workload is big, inspection cost is high, the efficiency is low, the colony lift assay is an effective, simple, convenient method and has great application value as a screening method in screening scFv antibody library of phage.
Acknowledgements
The authors are indebted to all the staff in the Chongqing Medical University School of Life Sciences, who provided assistance in the study.
Disclosure statement
The authors declare that they have no conflict of interest.
Additional information
Funding
References
- Rauchenberger R, Borges E, Thomassen-Wolf E, et al. Human combinatorial Fab library yielding specific and functional antibodies against the human fibroblast growth factor receptor 3. J Biol Chem. 2003;278:38194–38205.
- Dreher ML, Gherardi E, Skerra A, Milstein C. Colony assays for antibody fragments expressed in bacteria. J Immunol Methods. 1991;139:197–205.
- Rodenburg CM, Mernaugh R Fau-Bilbao G, et al. Production of a single chain anti-CEA antibody from the hybridoma cell line T84.66 using a modified colony-lift selection procedure to detect antigen-positive ScFv bacterial clones. Hybridoma. 1998;17:1–8.
- Radosević K, Voerman JS, Hemmes A, et al. Colony lift assay using cell-coated filters: a fast and efficient method to screen phage libraries for cell-binding clones. J Immunol Methods. 2003;272:219–233.
- Gao C, Mao S, Ronca F, et al. De novo identification of tumor-specific internalizing human antibody-receptor pairs by phage-display methods. J Immunol Methods. 2003;274:185–197.
- Chen RH, Liu QJ, Liao SQ. Research progress of medullary thyroid carcinoma. Gansu Med J. 2012;31:355–359.
- Mi YJ, Ma ZK, Zhao WJ. Progress for phage display technique and its application. Prog Mod Biomed. 2012;12:2766–2768.
- Gu YY, Yang XY, Zhang XK. Biochem sex lung cancer phage display antibody library construction. Guide China Med. 2012;10: 22–24.
- He J, Zhou G, Liu KD, Qin XY. Construction and preliminary screening of a human phage single-chain antibody library associated with gastric cancer. J Surg Res. 2002;102:150–155.
- Marks JD, Ouwehand WH, Bye JM, et al. Human antibody fragments specific for human blood group antigens from a phage display library. Biotechnology (N Y). 1993;11:1145–1149.
- Nie YZ, He Ft, Fau-Li ZK, et al. Identification of tumor associated single-chain Fv by panning and screening antibody phage library using tumor cells. World J Gastroenterol. 2002; 8:619–623.
- Hoogenboom HR, Lutgerink JT, Pelsers MM, et al. Selection-dominant and nonaccessible epitopes on cell-surface receptors revealed by cell-panning with a large phage antibody library. Eur J Biochem. 1999;260:774–784.
- Sanna PP, Williamson RA, De Logu A, et al. Directed selection of recombinant human monoclonal antibodies to herpes simplex virus glycoproteins from phage display libraries. Proc Natl Acad Sci U S A. 1995;92:6439–6443.
- Portolano S, McLachlan S, Rapoport B. High affinity, thyroid-specific human autoantibodies displayed on the surface of filamentous phage use V genes similar to other autoantibodies. J Immunol. 1993;151:2839–2851.
- Tur MK, Rothe A, Fau-Huhn M, et al. A novel approach for immunization, screening and characterization of selected scFv libraries using membrane fractions of tumor cells. Int J Mol Med. 2003;11:523–527.
- Zhou M, Yang DH, Tang SH, et al. Colony lift assay in the application of anti-liver cancer screening phage single antibody libraries. Chin J Pathol. 2005;21:1246–1248.
- Hu XL, Wang ZJ, Pang H. [Construction of human medullary thyroid carcinoma phage antibody library and preliminary identification]. J Chongqing Med Univ. 2013;38:1040–1043. Chinese.