Abstract
The most important role of molecular chaperones, especially under extreme temperature, is in handling with the impact of such stress on proteins and avoiding loss of their native conformation. Molecular chaperones play roles in the folding, transport, synthesis and quality control of other proteins under extreme temperature. The genome of the cyanobacterium Synechocystis PCC 6803 includes three dnaK genes (encoding heat-shock protein 70, Hsp70). The role of the second Hsp70 protein (Sll0170; DnaK2) in the acquisition of thermotolerance in Escherichia coli cells was investigated. Escherichia coli strain BL21 (DE3) was transformed with an expression vector (pTYB21) carrying the coding sequence of the Sll0170 gene under the control of the lac promoter. The recombinant cells which showed inducible expression of DnaK2 were observed to have improved viability compared to the wild-type strain upon transfer from 37 °C to 52 °C for up to 2, 4 and 6 min. Moreover, the growth of the wild-type cell culture decreased to 64% of that of the recombinant cell culture, following a transient (15 min) heat shock at 45 °C after which the culture was subsequently transferred back to 37 °C. Interestingly, the recombinant E. coli cells showed significantly faster culture growth than the wild-type cells at 37 °C.
Introduction
Cyanobacteria, or blue-green algae, are a group of photoautotrophs that originated about 3.5 billion years ago. Cyanobacteria are ubiquitous in distribution and are often found in extreme environmental conditions such as hot springs (Synechococcus sp., 70 °C; Oscillatoria terebriformis, ∼54 °C). They also exhibit the ability to survive in extremes of temperatures from 26 °C to 74 °C (Synechococcus lividus) or salinity stress (Aphanothece halophytica).[Citation1,Citation2] The cyanobacterium Synechocystis sp. PCC 6803 is a photosynthetic prokaryote which is believed to be a progenitor of higher plant chloroplasts. This organism has a relatively simple genome containing 3264 genes on the chromosome and its genome has been completely sequenced.[Citation3,Citation4] Cyanobacterial genomes typically encode multiple heat-shock protein 70 (Hsp70) (DnaK) chaperones, and in the genome of the cyanobacterium Synechocystis PCC 6803, three DnaK proteins are encoded.[Citation4,Citation5] Proteins of the 70 kDa heat-shock protein family (Hsp70) are ubiquitously distributed in all kingdoms of life, and in eukaryotes members of this family can be found in several organelles as well as in the cytosol.[Citation3,Citation6] The main functions of the proteins are to ensure proper folding of polypeptide chains, to keep proteins in a translocation-competent state after synthesis, and to prevent misfolding and aggregation of proteins caused by e.g. cellular stress.[Citation6]
Apparently, DnaK2 seems to be strictly conserved in all cyanobacterial lineages, and the phylogenetic analysis of DnaK indicates that it represents the only DnaK retained in algal and plant plastids.[Citation4,Citation7,Citation8] Complete genome sequencing revealed the presence of three DnaK homologues (DnaK1, DnaK2 and DnaK3) in Synechocystis PCC 6803.[Citation2,Citation8] Synechocystis DnaK2 is mainly detected in soluble protein and is also strongly bound to membranes peripherally.[Citation5–7] Previously, DNA microarray analysis has shown that the expression of DnaK2 in Synechocystis PCC 6803 is enhanced by high light, salt, heat, hyperosmotic, peroxide, ultraviolet (UV)-B radiation or acid condition (pH 3.0) stress.[Citation7,Citation8] Moreover, northern blot analysis has demonstrated that Synechocystis DnaK2 is induced by ethanol, heat, hydrogen peroxide, UV or salt stress.[Citation9,Citation10] Also, the level of Synechocystis DnaK2 protein is further enhanced following heat-shock treatment.[Citation10] Several studies indicate that DnaK2 is essential for the growth of Synechocystis under normal conditions.[Citation5,Citation6] Therefore, it could be suggested that, among Synechocystis DnaKs, DnaK2 is the most abundant protein and that DnaK2 has some specific functions that cannot be compensated for by the remaining two DnaK proteins.[Citation5,Citation6,Citation11] Transformation of heat-shock genes from Synechocystis sp. PCC 6803 into the bacterial host Escherichia coli cells is one of the most fundamental and indispensable techniques for studying gene functions. Ever since Mandel and Higa [Citation12] proposed the chemical transformation protocol in 1970, considerable variations in this basic technique have been studied.[Citation5,Citation13,Citation14]
The aim of this study was to isolate the Sll0170 gene (DnaK2) from Synechocystis PCC 6803 and validate its functions, using a gain-of-function approach by overproduction of DnaK2 in E. coli. The recombinant E. coli cells showed significant increased resistance to heat-shock and high-temperature stresses.
Materials and methods
Chemicals and reagents
BL21 (DE3) competent E. coli cells, ampicillin, 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal) and isopropyl-β-D-thiogalactopyranoside (IPTG) were purchased from UFC Biotechnology (USA). Restriction enzymes and pTVB21 expression vector were obtained from New England Biolab (UK). pGEM-T Easy cloning vector was purchased from Promega (USA). Other chemicals were purchased with the highest quality commercially available.
Bacterial strains and growth conditions
The wild-type strain Synechocystis sp. PCC 6803 was cultured photoautotrophically in Allen's medium [Citation1] at 27 °C for 5–7 days under illumination (20 µE S−1 m−2) with bubbling of sterile air. E. coli strain DH5α was used as a host cell for cloning, while E. coli strain BL21 (DE3) was used as a host for gene expression. Bacterial cultures were stored as 25% (v/v) glycerol stocks at −80 °C and maintained on Luria–Bertani (LB) plates containing 1.5% (w/v) agar. Cells harbouring recombinant plasmids were grown and maintained on LB media supplemented with 100 μg mL−1 ampicillin.
Expression of the Sll0170 gene in Escherichia coli
Chromosomal DNA was isolated from Synechocystis PCC 6803 by the method of Williams.[Citation15] The DNA fragment containing the open reading frame of the Sll0170 gene was amplified by polymerase chain reaction (PCR) with the following primers: 5′–GCG GCC GCC TAT TTC TCC GGC TCA GAG AAT T–3′ (Sll0170F) and 5′–CTC GAG ATG GGA AAA GTT GTT GGG AT–3′ (Sll0170R). The forward primer was designed to introduce a NotI site, while the reverse primer was designed to introduce an XhoI site (sequences in boldface). The amplified DNA fragment was cloned into a pGEM-T Easy vector (Promega, USA) and sequenced with an automated DNA sequencer (3130 Genetic Analyzer, Applied Biosystems, Japan). For the construction of the plasmid to express Sll0170, the pGEM/Sll0170 cloning vector was digested with NcoI and SalI. Next, the 1.9 kbp DNA fragment was cloned into the pTYB21 expression vector (New England Biolab, UK) digested with the same restriction enzyme. The resulting construct, designated pTYB21/Sll0170, was introduced into E. coli strain BL21 (DE3).
Production of recombinant protein in Escherichia coli and SDS-PAGE
Escherichia coli BL21 (DE3) transformed with pTYB21/Sll0170 gene was grown at 37 °C overnight in 5 mL LB medium supplemented with ampicillin at a final concentration of 100 µg mL−1. The cultures were then inoculated with a final volume 100 mL of LB culture. When the culture reached absorbance from 0.4 to 0.6 at 600 nm, 400 µmol L−1 IPTG was added and the bacteria were grown for further 3 h at 37 °C. Cells were harvested by centrifugation at 5000 r min−1 for 10 min at 4 °C and the pellets were kept frozen at −20 °C for analysis of the accumulation of the studied protein by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). SDS-PAGE was performed in 12% (w/v) polyacrylamide slab gels as previously described.[Citation16]
Response of Escherichia coli to high-temperature stress conditions
For the temperature stress experiment, overnight cultures of wild-type E. coli BL21 (DE3) cells with pTYB21 vector only and pTYB21/Sll0170 cells were grown in fresh LB liquid medium containing 100 μg ml−1 ampicillin under continuous shaking condition at 37 °C. When the absorbance at 600 nm (A600) reached a value of 0.2, the desired concentrations of 0.4 mmol L−1 IPTG were added and the samples were left for another 3 h. For heat-shock experiments, whole flasks were transferred to a second water bath operating at 45 °C and transferred back to 37 °C after 15 min. For high-temperature stress experiments, appropriate quantities of cell suspension grown in flasks were placed directly in the shaker water bath operating at 52 °C and left for up to 6 min and then cells were plated on LB agar medium (1.5%) and incubated overnight at 37 °C.
Database search and sequence evaluation
The genome of Synechocystis sp. PCC 6803 was searched for proteins representing DnaK2. The candidate protein of putative DnaK2 was verified by alignment with the protein sequence of other DnaKs from several organisms in ClustalW.[Citation17] The similarity per cent between different amino acid sequences was detected using the LALIGN program from the ExPASy tool. The protein size of DnaK2 as number of amino acids, molecular mass and pI value was calculated utilizing the Prot Param tools of the ExPASy-Proteomics Server (www.expasy.org/tools/protparam.html).
Results and discussion
Isolation and characterization of the Sll0170 gene
Heat stress disturbs cellular homeostasis and can lead to severe retardation in growth and development, and even death.[Citation18] Temperature stress can also have a devastating effect on cell metabolism, acting first on protein complexes, the quaternary structure being the first folding that is lost during heating. One of the side effects is the uncoupling of pathways, most of which involve electron transfer, resulting in transition of electrons in high-energy state, with concomitant formation of reactive oxygen species.[Citation17] Heat stress as well as other stresses can trigger some mechanisms of defence, such as the obvious gene expression that was not expressed under ‘normal’ conditions.[Citation6] For review see.[Citation19] In fact, this response to stresses on the molecular level is found in all living things, especially the sudden changes in gene expression resulting in an increase in the synthesis of certain protein groups known as Hsps.[Citation3,Citation18] Thermotolerance of cyanobacterial species, both unicellular and filamentous, is enhanced upon pre-treatment at sublethal temperatures, suggesting involvement of heat-shock genes/proteins in thermotolerance.[Citation6] Although some Hsp70 proteins are involved in the cellular heat-shock response, the proteins are usually constitutively expressed and functional, and therefore members of the Hsp70 family are better described as chaperones rather than as Hsps.[Citation6,Citation19] After annotation of the first completely sequenced genome of a cyanobacterium, it became evident that three proteins are encoded by dnaK gene families in Synechocystis sp. PCC 6803.[Citation2,Citation6]
In this study, we isolated and characterized the gene (Sll0170) encoding Hsp70 protein (DnaK2) from Synechocystis PCC6803. The Sll0170 gene was shown to consist of 1911 bp encoding 636 amino acids with a calculated molecular mass of 67.614 kDa and a pI of 4.72. When the PCR product of the Sll0170 gene fragment (1911 bp) was sequenced with an automated DNA sequencer, computer-assisted DNA alignments suggested that a large part of a gene homologous to the Sll0170 gene in Synechocystis PCC 6803 (encoding the DnaK2 protein [Citation2]) was present within the 1911 bp cloned fragment. Members of the Hsp70 family are highly conserved molecular chaperones that are found in eubacteria, archaebacteria and eukaryotes.[Citation18] A comparison of the deduced amino acid sequence of the DnaK2 protein from Synechocystis PCC 6803 with the sequences of DnaK2 proteins from prokaryotic algae, yeasts, higher plants and human are shown in . The deduced amino acid sequence of DnaK2 from Synechocystis PCC 6803 shared 27.9%–97.8% identity to those of DnaK2 proteins from human, higher plants, yeast, bacteria and Synechocystis PCC 6714 (). The predicted sequence of the DnaK2 protein showed the highest sequence similarity to the homologues in prokaryotic algae, bacterial DnaK proteins as well as to higher plants DnaK2 proteins ( and ). The putative amino acid sequence of Synechocystis DnaK2 is highly conserved with other DnaK2 proteins from other organisms especially in the N-terminus, which contains the ATPase domain (1–385 a.a.) and substrate domain (386–513 a.a.).[Citation20] These results are in agreement with the research reported by Rupprecht et al.,[Citation10] who found that Synechocystis DnaK2 protein shows the highest sequence identity to bacterial DnaK proteins as well as to plant chloroplast Hsp70 proteins. The observed differences were mainly in the C-terminal extensions of the protein (514–638 a.a.) (). The C-terminal region does not show a high degree of sequence similarity, but the C-terminal domains of Hsp70 proteins are in general not highly conserved.[Citation10]
Figure 1. Comparison of a part of the deduced amino acid (aa) sequence of Synechocystis PCC 6803 DnaK2 (636 aa) with those of DnaK2 proteins from Synechocystis PCC 6714 (637 aa), Staphylococcus aureus (610 aa), Saccharomyces cervisiae (657 aa), Oryza sativa Indica Group (648 aa), Arabidopsis thaliana (650 aa) and Homo sapiens (701 aa), using the single-letter code (CLUSTALW).
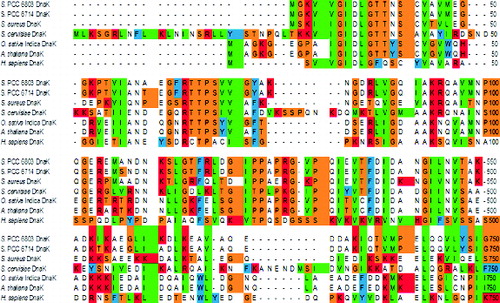
Table 1. Identity and similarity per cent of Synechocystis PCC 6803 DnaK2 protein with DnaK proteins from some prokaryotic and eukaryotic organisms (LALIGN).
Expression of the Sll0170 gene in Escherichia coli
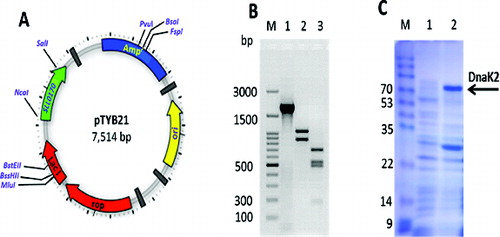
The availability of the full-length DNA encoding Sll0170 enabled us to amplify, by PCR, a fragment of this clone for the production of the mature form of the DnaK2 protein in E. coli cells (). The NotI and XhoI sites included in the PCR products were used to clone the fragment into the expression vector pTYB21 ((A)). No expression was detectable when the PCR product of the Sll0170 gene cloned fragment was in the opposite orientation with respect to the PTYB21 lac promoter, suggesting that the Sll0170 gene promoter was recognized poorly or not at all by E. coli RNA polymerase (data not shown). The resulting construct designated pTYB21/Sll0170 was used to transform E. coli strain BL21 (DE3) cells. To further check the full DNA sequence of the Sll0170 gene, a restriction map for the gene was done by cutting the gene with XbaI ((B), lane 2) and SmaI ((B), lane 3). Next, we examined the optimum conditions for the expression of Sll0170 that was sub-cloned into the pTYB21 expression vector. After induction with IPTG, the recombinant enzyme was expressed at a high level in E. coli cells. As shown in (C), the protein band corresponding to the recombinant DnaK2, which correlated with a molecular mass of 67.614 kDa calculated from the deduced amino acid sequence, was observed. The recombinant DnaK2 protein accounted for nearly 30% of the total protein in the E. coli cells ((C), lane 2). There was no protein band corresponding to DnaK2 in the control cells ((C), lane 1). Interestingly, a 32 kDa band was more highly expressed in recombinant cells than in wild-type cells ((C)). This band could correspond to transcription factor σ32, as Liberek et al. [Citation21] reported that in E. coil cells, the heat-shock response is positively regulated by the σ32 transcriptional factor and during the purification procedure of this factor a large fraction of the overexpressed σ32 polypeptide copurified with the universally conserved DnaK Hsp (the prokaryotic equivalent of the Hsp70). Thus, they confirmed that purified σ32 bound to DnaK.[Citation21]
Figure 2. Expression of the Sll0170 gene in E. coli. (A) Construction of the plasmid pTYB21/Sll0170. (B) PCR amplification of the Sll0170 gene from Synechocystis PCC 6803 (lane 1) and restriction map of the Sll0170 gene using XbaI (lane 2) and SmaI (lane 3), while lane M is 100 bp DNA ladder (Promega). (C) SDS-PAGE analysis of the recombinant DnaK2 expressed in E.coli cells. Lane 1, pTYB21-only transformed E. coli; lane 2, pTYB21/Sll0170-transformed E. coli; lane M, Page Ruler Prestained Protein Ladder (Life Technologies).
Effect of recombinant DnaK2 on tolerance to heat-shock and high-temperature stresses in Escherichia coli cells
While the exact physiological functions of multiple DnaK proteins in cyanobacteria are still unknown, only DnaK2 and DnaK3 are essential in Synechocystis and Synechococcus.[Citation5,Citation6,Citation22] Accordingly, DnaK2 seems to be the major Hsp70 protein involved in the stress response,[Citation6] whereas DnaK3 has a specialized function, which involves the highly conserved prolonged C-terminus of the protein.[Citation22] In contrast to other bacteria, neither dnaK nor any other heat-shock genes such as grpE and dnaJ are located near the hrcA genes in Synechocystis and other cyanobacteria.[Citation7,Citation23,Citation24] It has been suggested that dnaK2 is unique among the three dnaK genes exhibiting a stress response, although they are expressed in the same cellular compartment.[Citation25] This suggestion strongly supports the functional assignment of these DnaK proteins, and the highly conserved sequence of DnaK2 implies that it has the most essential Hsp70 functions.[Citation25]
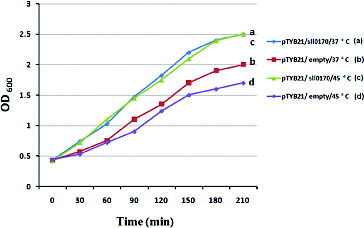
In this study, E. coli was used as a model system to test whether the recombinant DnaK2 can protect the microorganism cells against high-temperature stress. Such stress was produced in vitro by transferring the cell cultures from 37 °C to 45 °C (heat-shock stress) for 15 min and then back to 37 °C. The cells were also exposed to high-temperature stress (52 °C) for different lengths of time. The macroscopic effects of growth perturbations are most easily seen as changes in growth rate.[Citation26] For this purpose, the growth of recombinant E. coli cells was compared to the growth of the wild-type strain in experiments, where E. coli BL21 (DE3) was subjected to temporary heat-shock stress (45 °C) (). Transient heat shock resulted in a significant decrease in the growth of wild-type cells compared to that of the recombinant cells. The recombinant cell culture reached higher optical density than wild-type cells both under prolonged exposure at 37 °C and under transient heat-shock exposure (45 °C for 15 min).
Figure 3. Growth of E. coli wild-type cells with an empty pTYB21 vector and recombinant cells with pTYB21/sll0170 in LB medium exposed to temperature shock. Growth curves are shown for two cultures. At time zero, one culture was maintained at 37 °C ((a) blue and (b) red line), while a second culture was transferred to 45 °C and then returned to 37 °C after 15 min ((c) green and (d) purple line). The culture density at time zero was OD600 = 0.4.
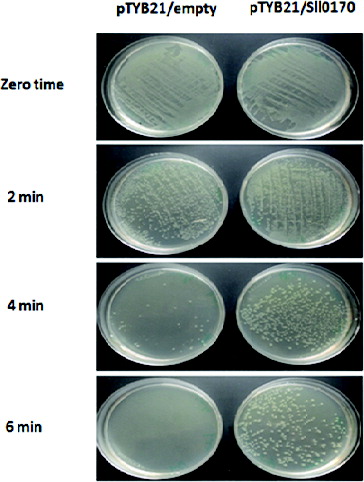
Next, in order to evaluate the protective efficiency of DnaK2 on cell growth under high-temperature stress, E. coli wild-type and recombinant cells were exposed to 52 °C for up to 6 min (). Interestingly, the growth of wild-type cells was severely reduced at 4 min and was obliterated at 6 min. On the other hand, the recombinant pTYB21/Sll0170 could rescue the growth of the E. coli strain BL21 (DE3) on the plate even when the cells were exposed to 52 °C for up to 6 min. All these results indicate that the overproduction of Synechocystis PCC 6803 Sll0170 gene in E. coli cells can improve their tolerance to heat-shock and high-temperature stress.
Figure 4. Effect of high-temperature stress (52 °C for 2, 4 and 6 min) on the growth of E. coli wild-type (pTYB21/empty) and recombinant (pTYB21/Sll0170) cells.
Survival of E. coli strains at high temperatures has been well studied and multiple dnaK genes have been reported.[Citation21] In E. coli, the dnaK gene is not essential for growth, but deletion of the gene confers a high-temperature sensitive phenotype.[Citation27] In general, of the three dnaK genes in the unicellular cyanobacteria, only dnaK2 is induced under heat and other abiotic stresses [Citation10,Citation23] and contributes to thermotolerance.[Citation10] For review see [Citation6,Citation28,Citation29]. For example, in Synechococcus elongatus PCC 7942, only the DnaK2 gene responds to heat, high light intensity, high salt concentration and osmotic stresses.[Citation8] As highlighted by Nakamoto,[Citation28] Synechococcus DnaK2 can suppress the temperature sensitive growth of an E. coli DnaK mutant strain at 44.5 °C, suggesting that DnaK2 is involved in cellular thermotolerance.[Citation11] On the other hand, the overproduction of DnaK1 and DnaK3 resulted in growth inhibition even at the permissive temperature 42 °C.[Citation11] In contrast to Synechococcus, none of the Synechocystis dnaK genes can complement the defect of the E. coli DnaK mutant.[Citation6] On the contrary to the latter, our data in and confirmed that the overproduction of Synechocystis DnaK2 can protect E. coli cells from high-temperature and heat-shock temperature stresses. It could be speculated that, under heat stress, DnaK2 may protect the cellular proteins from misfolding and aggregation and thus keep the proteins in a translocation state after synthesis. This may explain why, among Synechocystis DnaKs, DnaK2 is the most abundant protein [Citation10] and the level of Synechocystis DnaK2 protein is further enhanced following heat-shock treatment.[Citation30]
Conclusions
This work describes cloning of the DnaK2 gene (Sll0170) of Synechocystis sp. PCC 6803 and overexpression of the protein in the cytoplasm of E. coli. The functions of the recombinant protein in E. coli cells were confirmed by SDS-PAGE and visible survival test. The results indicate that Synechocystis DnaK2 confers tolerance to high-temperature and heat-shock stress in E. coli, thus, confirming absence of species barrier in terms of the DnaK2 functioning.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Whitton BA, Potts M. The ecology of cyanobacteria: their diversity in time and space. Dordrecht: Kluwer Academic Publisher; 2000.
- Zhang Y, Niu X, Shi M, et al. Identification of a transporter Slr0982 involved in ethanol tolerance in cyanobacterium Synechocystis sp. PCC 6803. Front Microbiol. 2015;6:1–13.
- Suzuki I, Simon WJ, Slabas AR. The heat shock response of Synechocystis sp. PCC 6803 analysed by transcriptomics and proteomics. J Exp Bot. 2006;57:1573–1578.
- Düppre E, Rupprecht E, Schneider D. Specific and promiscuous functions of multiple DnaJ proteins in Synechocystis sp. PCC 6803. Microbiology. 2011;157:1269–1278.
- Kim J, Ahn M, Park Y, et al. Synechocystis PCC6803 and PCC6906 dnaK2 expression confers salt and oxidative stress tolerance in Arabidopsis via reduction of hydrogen peroxide accumulation. Mol Biol Rep. 2014;41:1091–1101.
- Rupprecht E, Gathmann S, Fuhrmann E, et al. Three different DnaK proteins are functionally expressed in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology. 2007;153:1828–1841.
- Nakamoto H, Suzuki M, Kojima K. Targeted inactivation of the hrcA repressor gene in cyanobacteria. FEBS Lett. 2003;549:57–62.
- Sato M, Nimura-Matsune K, Watanabe S, et al. Expression analysis of multiple dnaK genes in the cyanobacterium Synechococcus elongatus PCC 7942. J Bacteriol. 2007;189:3751–3758.
- Nakamoto H, Fujita K, Ohtaki A, et al. Physical interaction between bacterial heat shock protein (Hsp) 90 and Hsp70 chaperones mediates their cooperative action to refold denatured proteins. J Biol Chem. 2014;289:6110–6119.
- Rupprecht E, Düppre E, Schneider D. Similarities and singularities of three DnaK proteins from the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2010;51:1210–1218.
- Nimura K, Takahashi H, Yoshikawa H. Characterization of the dnaK multigene family in the cyanobacterium Synechococcus sp. strain PCC7942. J Bacteriol. 2001;183(4):1320–1328.
- Mandel M, Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970;53:159–162.
- Fregel R, Rodriguez V, Cabrera VM. Microwave improved Escherichia coli transformation. Lett Appl Microbiol. 2008;46:498–499.
- Aune TEV, Aachmann FL. Methodologies to increase the transformation efficiencies and the range of bacteria that can be transformed. Appl Microbiol Biotechnol. 2010;85:1301–1313.
- Williams JGK. Construction of specific mutants in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol. 1988;167:766–778.
- Laemmli UK. Cleavage of structural proteins during the assembly of head of Bacteriophage T4. Nature. 1970;227:680–685.
- Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680.
- Zhou W, Zhou T, Li M-X, et al. The Arabidopsis J-protein AtDjB1 facilitates thermotolerance by protecting cells against heat-induced oxidative damage. New Phytologist. 2012;194:364–378.
- Al-Whaibi MH. Plant heat-shock proteins: a mini review. J King Saud Univ – Sci. 2011;23:139–150.
- Itoh T, Matsuda H, Mori H. Phylogenetic analysis of the third Hsp70 homolog in Escherichia coli; a novel member of the Hsc66 subfamily and its possible co-chaperone. DNA Res. 1999;6:299–305.
- Liberek K, Galitski TP, Zylicz M, et al. The DnaK chaperone modulates the heat shock response of Escherichia coli by binding to the σ32 transcription factor. Proc Natl Acad Sci USA. 1992;89:3516–3520.
- Horváth I, Glatz A, Nakamoto H, et al. Heat shock response in photosynthetic organisms: membrane and lipid connections. Prog Lipid Res. 2012;51:208–220.
- Genevaux P, Georgopoulos C, Kelley WL. The Hsp70 chaperone machines of Escherichia coli: a paradigm for the repartition of chaperone functions. Mol Microbiol. 2007;66:840–857.
- Hinault MP, Cuendet AFH, Mattoo RUH, et al. Stable β-synuclein oligomers strongly inhibit chaperone activity of the Hsp70 system by weak interactions with J-domain co-chaperones. J Biol Chem. 2010;285:38173–38182.
- Dörrich AK, Mitschke J, Siadat O, et al. Deletion of the Synechocystis sp. PCC 6803 kaiAB1C1 gene cluster causes impaired cell growth under light–dark conditions. Microbiology. 2014;160:2538–2550.
- Mason CA, Dunner J, Indra P, et al. Heat-induced expression and chemically induced expression of the Escherichia coli stress protein HtpG are affected by the growth environment. Appl Environ Microbiol. 1999;3433–3440.
- Paek KH, Walker GC. Escherichia coli dnaK null mutants are inviable at high temperature. J Bacteriol. 1987;169:283–290.
- Nakamoto H. Molecular chaperones and stress tolerance in cyanobacteria. In: Srivastava AK, Rai AN, Neilan BA, editors. Stress biology of cyanobacteria: molecular mechanisms to cellular responses. Boca Raton (FL): CRC Press; 2013. p. 114–144.
- Rajaram H, Chaurasia A, Apte S. Cyanobacterial heat-shock response: role and regulation of molecular chaperones. Microbiology. 2014;160:647–658.
- Bracher A, Verghese J. GrpE, Hsp110/Grp170, HspBP1/Sil1 and BAG domain proteins: nucleotide exchange factors for Hsp70 molecular chaperones. Sub-cellular Biochem. 2015;78:1–33.
